Living Microbes Discovered In Earth’s Driest Desert
The Atacama Desert, which runs along the Pacific Coast in Chile, is the driest place on the planet and, largely because of that aridity, hostile to most living things. Not everything, though—studies of the sandy soil have turned up diverse microbial communities. Studying the function of microorganisms in such habitats is challenging, however, because it’s difficult to separate genetic material from the living part of the community from genetic material of the dead.
A new separation technique can help researchers focus on the living part of the community. This week in Applied and Environmental Microbiology, an international team of researchers describes a new way to separate extracellular (eDNA) from intracellular (iDNA) genetic material. The method provides better insights into microbial life in low-biomass environments, which was previously not possible with conventional DNA extraction methods, said Dirk Wagner, Ph.D., a geomicrobiologist at the GFZ German Research Centre for Geosciences in Potsdam, who led the study.

Location of the study sites and bacterial abundances. (A) Location of the study sites along the Atacama Desert. Moisture gradient including Coastal Sand (CS), Alluvial Fan (AL), Red Sands (RS), Yungay (YU), and two hyperarid reference sites, Maria Elena (ME) and Lomas Bayas (LB). — Applied and Environmental Microbiology
The microbiologists used the novel approach on Atacama soil samples collected from the desert along a west-to-east swath from the ocean’s edge to the foothills of the Andes mountains. Their analyses revealed a variety of living and possibly active microbes in the most arid areas. A better understanding of eDNA and iDNA, Wagner said, can help researchers probe all microbial processes.
“Microbes are the pioneers colonizing this kind of environment and preparing the ground for the next succession of life,” Wagner said. These processes, he said, aren’t limited to the desert. “This could also apply to new terrain that forms after earthquakes or landslides where you have more or less the same situation, a mineral or rock-based substrate.”
Most commercially available tools for extracting DNA from soils leave a mixture of living, dormant and dead cells from microorganisms, Wagner said. “If you extract all the DNA, you have DNA from living organisms and also DNA that can represent organisms that just died or that died a long time ago.” Metagenomic sequencing of that DNA can reveal specific microbes and microbial processes. However, it requires sufficient good-quality DNA, Wagner added, “which is often the bottleneck in low-biomass environments.”
To remedy that problem, he and his collaborators developed a process for filtering intact cells out of a mixture, leaving behind eDNA genetic fragments left from dead cells in the sediment. It involves multiple cycles of gentle rinsing, he said. In lab tests they found that after 4 repetitions, nearly all the DNA in a sample had been divided into the 2 groups.
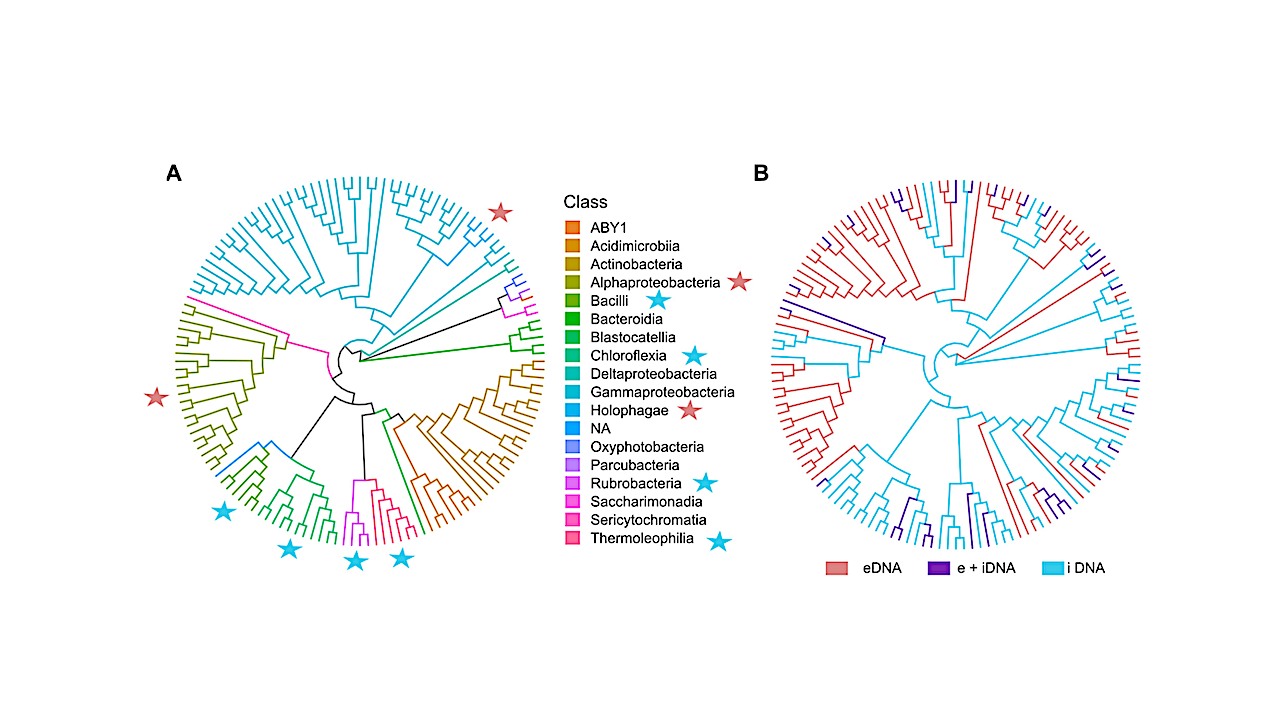
When they tested soil from the Atacama Desert, they found Actinobacteria and Proteobacteria in all samples in both eDNA and iDNA groups. That’s not surprising, Wagner said, because the living cells constantly replenish the store of iDNA as they die and degrade. “If a community is really active, then a constant turnover is taking place, and that means the 2 pools should be more similar to each other,” he said. In samples collected from depths of less than 5 centimeters, they found that Chloroflexota bacteria dominated in the iDNA group.
In future work, Wagner said he plans to conduct metagenomic sequencing on the iDNA samples to better understand the microbes at work, and to apply the same approach to samples from other hostile environments. By studying iDNA, he said, “you can get deeper insights into the real active part of the community.”

Relative abundances of the top 161 ASVs (>0.1% mean abundance) summarized by family level for the e- and iDNA pools along the moisture gradient: Coastal Sand (CS), Aluvial Fan (AL), Red Sands (RS), Yungay (YU), and additional hyperarid study sites Lomas Bayas (LB) and Maria Elena (ME). Bubbles represent the mean value of relative abundances from three technical replicates. ASVs are sorted by phylum and characterized by their family name; otherwise, the next highest classification level was chosen (o = order, c = class). k_Bacteria represents an unknown environmental clone related to Firmicutes (86% similarity) and Chloroflexi (87% similarity). — Applied and Environmental Microbiology
Inside the Atacama Desert: uncovering the living microbiome of an extreme environment, Microbial Ecology (open access)
Astrobiology