Ocean World Genomics: Molecular Monitoring For Marine Change And Threats Using eDNA

New DNA* technology promises to improve efforts to monitor marine life in the Southern Ocean, and detect the presence of non-native marine species close to Antarctica.
During a 3,000 nautical mile voyage from Hobart to Davis research station in 2019, on former Australian icebreaker Aurora Australis, scientists collected 138 seawater samples and examined the ‘environmental DNA’ (eDNA) in the samples to see which zooplankton species (tiny marine animals) had been present at the time the samples were collected.
Australian Antarctic Division molecular geneticist, Dr Leonie Suter, said they then compared the species detected using eDNA, to zooplankton collected using a continuous plankton recorder (CPR).
“The CPR is an instrument towed behind the ship that captures zooplankton in between two sheets of silk,” Dr Suter said.
“We can then look at the captured animals under the microscope to identify them.”
The CPR has been used to monitor Southern Ocean zooplankton diversity since 1991, providing a valuable long-term data set that allows scientists to see changes in zooplankton communities.
However, only more robust organisms, such as crustaceans, remain intact in the CPR, while many fragile and gelatinous organisms get damaged and cannot be identified.
“With eDNA we can identify most things in the environment, even if it’s only a tiny part of it, or faeces, eggs, or larvae,” Dr Suter said.
“So we wanted to compare the information eDNA can provide, with the CPR, to see if eDNA can make a useful contribution to the existing Southern Ocean zooplankton data set.”
The team, led by former Curtin University researcher Georgia Nester (now at Mindaroo Foundation), found that eDNA detected a wider range of species than the CPR, and could be used in sea ice, where the CPR cannot go.
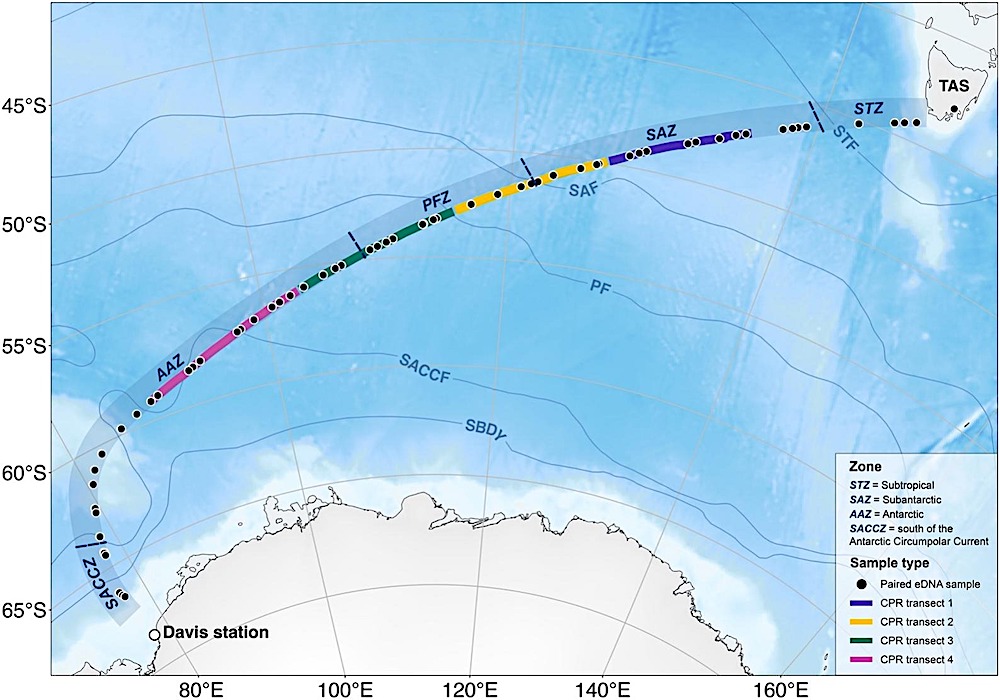
Map of the Southern Ocean sampling region with sampling conducted from north to south. Transects began at Hobart, Tasmania, Australia (TAS) and ended at Davis Station, Antarctica. eDNA samples are indicated by the black dots, and CPR transects by coloured lines: transect 1 (purple), 2 (yellow), 3 (green), and 4 (pink) differentiated. Fronts are visualised according to Orsi et al. (1995) and described in Section 2.5 . Mean sea-ice extent for November 2019 is indicated by the white line using data from the National Snow and Ice Data Center ( Fetterer et al., 2017 ). Map created using “SOmap” R package ( Maschette et al., 2019 ).
“We detected 68 zooplankton species using eDNA and 32 using CPR tows,” Dr Nester said.
“Only 12 species were shared between the two methods and most of these were crustaceans, such as krill and copepods.”
Dr Suter said eDNA and CPR methods had different strengths and limitations, so the combination of both methods would “widen the lens”.
“The two methods together provide a more holistic view of zooplankton diversity and change, and may allow us to see patterns we might otherwise miss,” Dr Suter said.
“For example, some gelatinous organisms may cope better with warming ocean temperatures, but if we’re only looking at crustaceans, we may miss that.”
During the voyage, the eDNA method also detected 16 non-native or potentially invasive species.
“Most of these species were detected closer to Tasmania where they are known to occur,” Dr Nester said.
“However, detections for some species increased once we entered the sea ice.
“These detections were largely attributed to organisms attached to the ship’s hull being abraded by the sea ice.
“While hull biofouling is a recognised potential pathway for marine introductions into Antarctica, overall eDNA detections were relatively low, and we cannot rule out that they represent non-viable DNA, larvae or dead organisms.”
Australian Antarctic Division Environmental Manager, Kirsten Leggett, said Australia’s new icebreaker, RSV Nuyina, has a biofouling management plan to minimse the risk of transporting non-native species to Antarctica, and complies with the International Maritime Organisation’s biofouling guidelines.
“We continue to engage with relevant authorities and subject matter experts to ensure the highest standards of biosecurity are achieved across the Australian Antarctic Program,” Ms Leggett said.
As well as being useful for monitoring long-term change, Dr Suter said eDNA provides a novel means of monitoring marine biosecurity risks.
“Given the vulnerability of Antarctic environments to the introduction of non-native species, future work should look at ways to increase the likelihood of detecting non-native species in Antarctica in a timely manner, and eDNA-based methods could be suitable,” she said.
“In addition, eDNA in combination with CPR surveys and biophysical data will proved a nuanced picture of Southern Ocean ecosystems, to better understand the impacts of changing conditions on these ecosystems.”
The research is published in Science of the Total Environment.
DNA is genetic information found in the cells of every organism. Organisms shed their DNA into the environment (from skin, scales and faeces, for example), where it gets mixed into a big ‘soup’ of ‘environmental DNA’, or eDNA – the DNA of all organisms present in that environment.
Astrobiology








