New Study Sheds Light on Ancient Protoribosome and its Role in Early Life Evolution
Scientists have uncovered new insights into the protoribosome, a molecular fossil within the ribosome, which plays a key role in the origin of life on Earth. This research, published in Nucleic Acids Research, could reshape our understanding of early ribosomal evolution and its significance for the development of life.
[This is a Joint Press Release with the Charles University, University of Chemistry and Technology in Prague, and University of Milano]
The study’s research team includes Dr. Klára Hlouchová from Department of Cell Biology of the Faculty of Science, Charles University and Associate Professor, Dr. Kosuke Fujishima of the Earth-Life Science Institute at the Institute of Science Tokyo (formerly Tokyo Institute of Technology).
Life as we know it nowadays relies on the intimate connection among nucleic acids, storing information, proteins, performing countless tasks, and lipids forming surrounding membranes. “These interactions among molecular precursors started to occur more than 4 billion years ago before the first life emerged,” says Dr. Klára Hlouchová from the Charles University, one of the study’s lead researchers.
Cells contain ribosomes, molecular machines which produce proteins. Due to their omnipresence and high conservation across all life forms, ribosomes qualify as ancient molecular fossils, attracting evolutionary biologists as the best connection to our biological past.
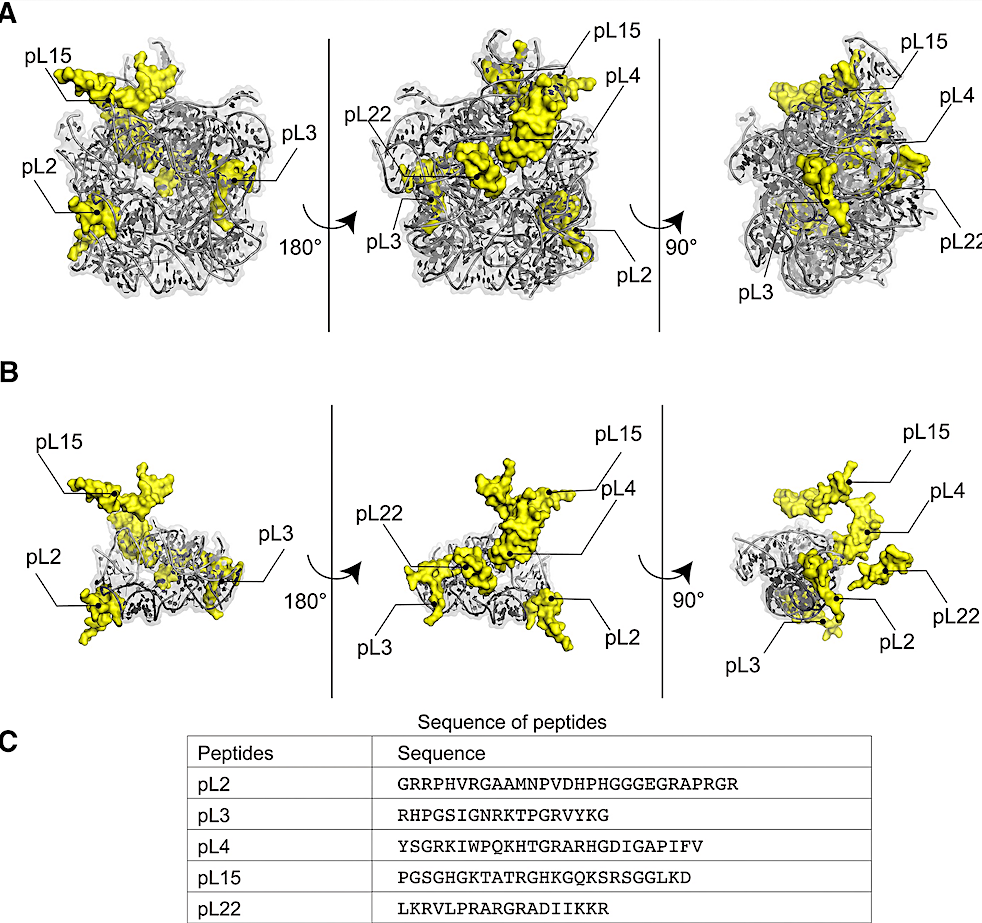
PTC models and rPeptide sequences. (A and B) The models of WT bPTC (A) and WT sPTC (B), represented as ribbon, and rPeptides (pL2, 3, 4, 15, 22). Both models are extracted from the Thermus thermophilus ribosomal structure (PDB ID: 4V51), missing the loops that were added by authors of the designed models (9,11). The two symmetrical A and P regions of the WT sPTC (B) are depicted with lighter and darker shades of grey, respectively. (C) Sequences of rPeptides used in this study. — Nucleic Acids Research
Key discovery
The protoribosome surrounds the peptidyl transferase center (PTC), which is responsible for peptide bond formation—an essential process in protein synthesis. Previous studies showed that RNA alone could exhibit PTC activity. However, in the ribosomal structure, “tails” of several ribosomal proteins (rPeptides) are located in proximity of the PTC and have been implied as relics of the most ancient peptide species that probably interacted with the protoribosome before the ribosome evolved into the RNA-protein complex as we know it today. The role of these rPeptides has not been studied so far.
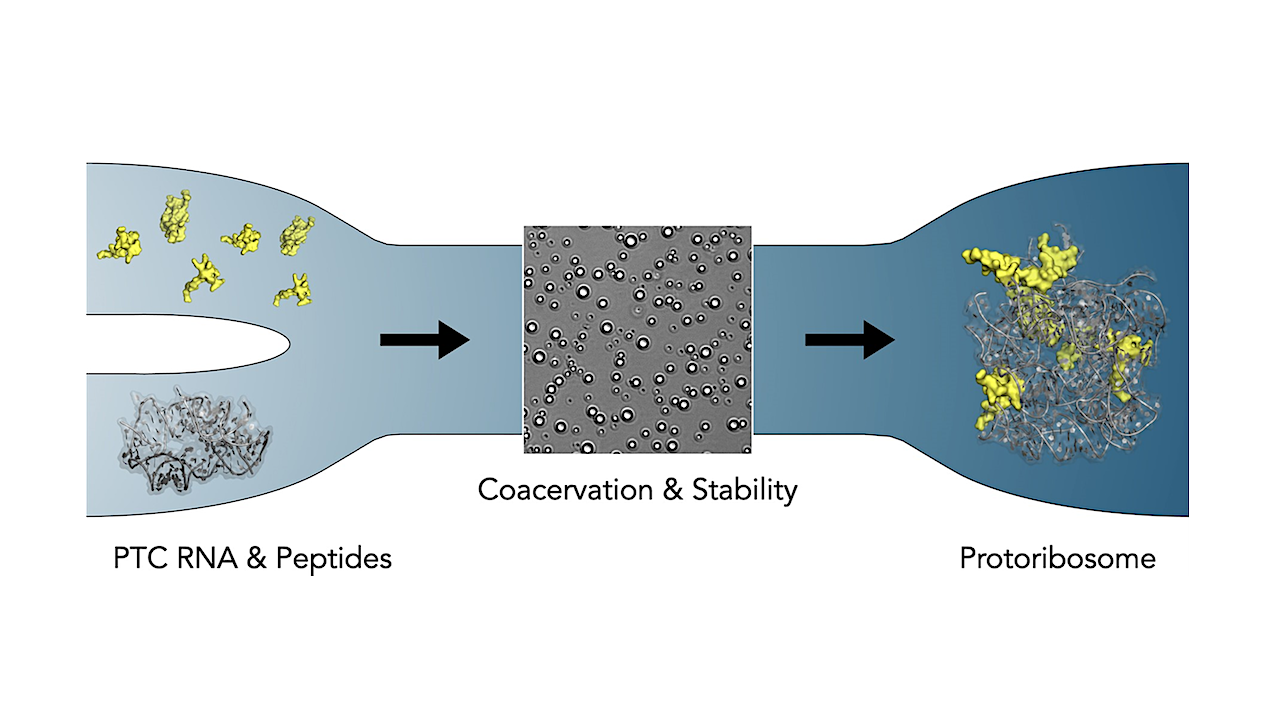
This new investigation reveals that the rPeptides have a critical role in driving compartmentalization and hence stability of the protoribosome. Two distinct evolutionary stages of the protoribosomal RNA have been studied here – The 617- and 136-nucleotide (“big” and “small”) constructs. The small construct (linked to the previously shown PTC activity) is more structurally flexible and while it interacts with rPeptides with lower specificity, the peptides induce coacervation—a process that helps form liquid-like droplets—across a wide concentration range.
This, in turn, protects RNA from degradation. The big construct, on the other hand, is structurally more defined, as shown by atomistic computer simulations performed by the group of associate professor Michal H. Kolář at University of Chemistry and Technology in Prague. The protoribosomal RNA in the big construct interacts with the rPeptides with higher specificity and coacervation of this complex is less prominent than of the small construct.
The distinct properties of the two protoribosomal stages suggest that rPeptides initially provided compartmentalization and prevented RNA degradation, preceding the emergence of specific RNA-protein interactions crucial for the structural integrity of the contemporary ribosome.
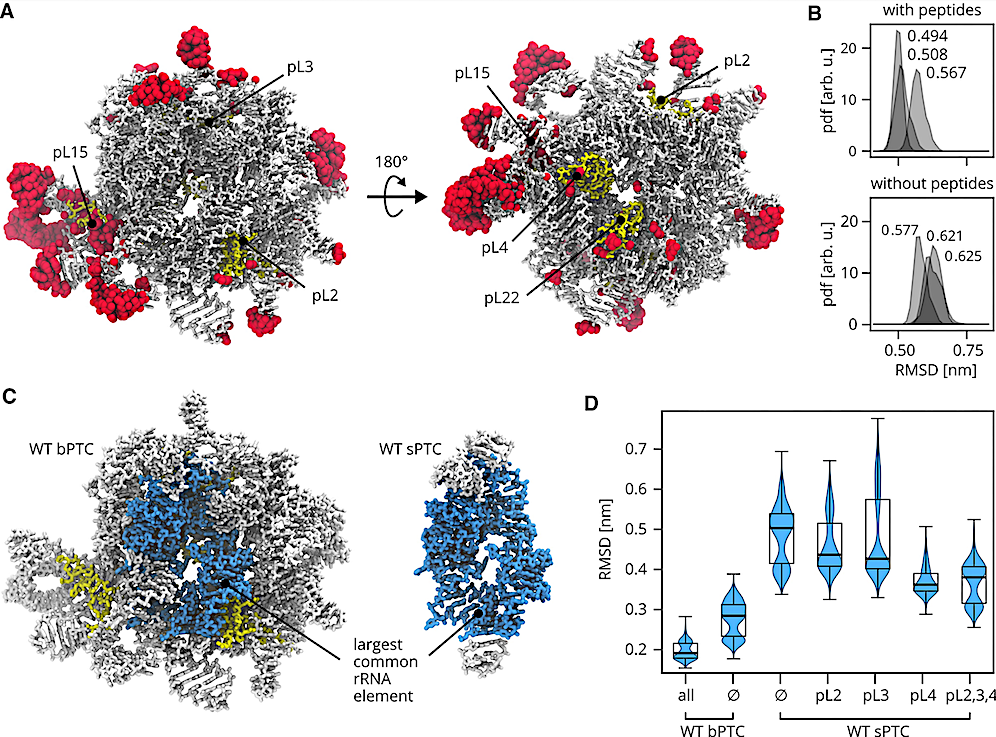
Overview of MD simulations. (A) The most flexible residues as identified by RMSF analysis of the last 800 ns of each trajectory. Top 10% residues with the highest RMSF are shown in surface representation, rRNA and rPeptides in stick representation. The two viewpoints show the tRNA-binding site and the ancestral exit tunnel lined by pL4 and pL22. For clarity, the location of pL3 is shown, although it is buried in the rRNA. (B) Probability density functions (pdfs) of RMSDs calculated for WT bPTC phosphorus atoms with respect to the initial conformation. The last 300 ns of each independent trajectory were used. Mean values of pdfs are denoted. (C) The largest common rRNA element (dark) in the context of WT bPTC and WT sPTC. (D) Violin plots of RMSD of the largest common rRNA element phosphorus atoms. The last 300 ns of each independent trajectory were used. Median of pdfs are denoted by the thick line, the box extends form the first to the third quartile, the whiskers extend from the box to the farthest data point lying within 1.5× the inter-quartile range. — Nucleic Acids Research
Evolutionary Implications
The research suggests that the interaction between RNA and proteins before the first life emerged, offered a significant biophysical advantage, especially by providing compartmentalization and preventing RNA from degradation. These early RNA-protein interactions are seen as a precursor to the more complex RNA-protein relationships that are essential for ribosomal structural integrity today.
The research was funded by Human Frontiers Science Foundation and the 4EU+ European University Alliance.
The interplay between peptides and RNA is critical for protoribosome compartmentalization and stability, Nucleic Acids Research (open access)
Abstract
The ribosome, owing to its exceptional conservation, harbours a remarkable molecular fossil known as the protoribosome. It surrounds the peptidyl transferase center (PTC), responsible for peptide bond formation. While previous studies have demonstrated the PTC activity in RNA alone, our investigation reveals the intricate roles of the ribosomal protein fragments (rPeptides) within the ribosomal core. This research highlights the significance of rPeptides in stability and coacervation of two distinct protoribosomal evolutionary stages. The 617nt ‘big’ protoribosome model, which associates with rPeptides specifically, exhibits a structurally defined and rigid nature, further stabilized by the peptides. In contrast, the 136nt ‘small’ model, previously linked to peptidyltransferase activity, displays greater structural flexibility. While this construct interacts with rPeptides with lower specificity, they induce coacervation of the ‘small’ protoribosome across a wide concentration range, which is concomitantly dependent on the RNA sequence and structure. Moreover, these conditions protect RNA from degradation. This phenomenon suggests a significant evolutionary advantage in the RNA–protein interaction at the early stages of ribosome evolution. The distinct properties of the two protoribosomal stages suggest that rPeptides initially provided compartmentalization and prevented RNA degradation, preceding the emergence of specific RNA–protein interactions crucial for the ribosomal structural integrity.
Astrobiology, genomics,