Immunoanalytical Detection of Conserved Peptides: Refining the Universe of Biomarker Targets in Planetary Exploration
Ancient peptides are remnants of early biochemistry that continue to play pivotal roles in current proteins. They are simple molecules yet complex enough to exhibit independent functions, being products of an evolved biochemistry at the interface of life and nonlife.
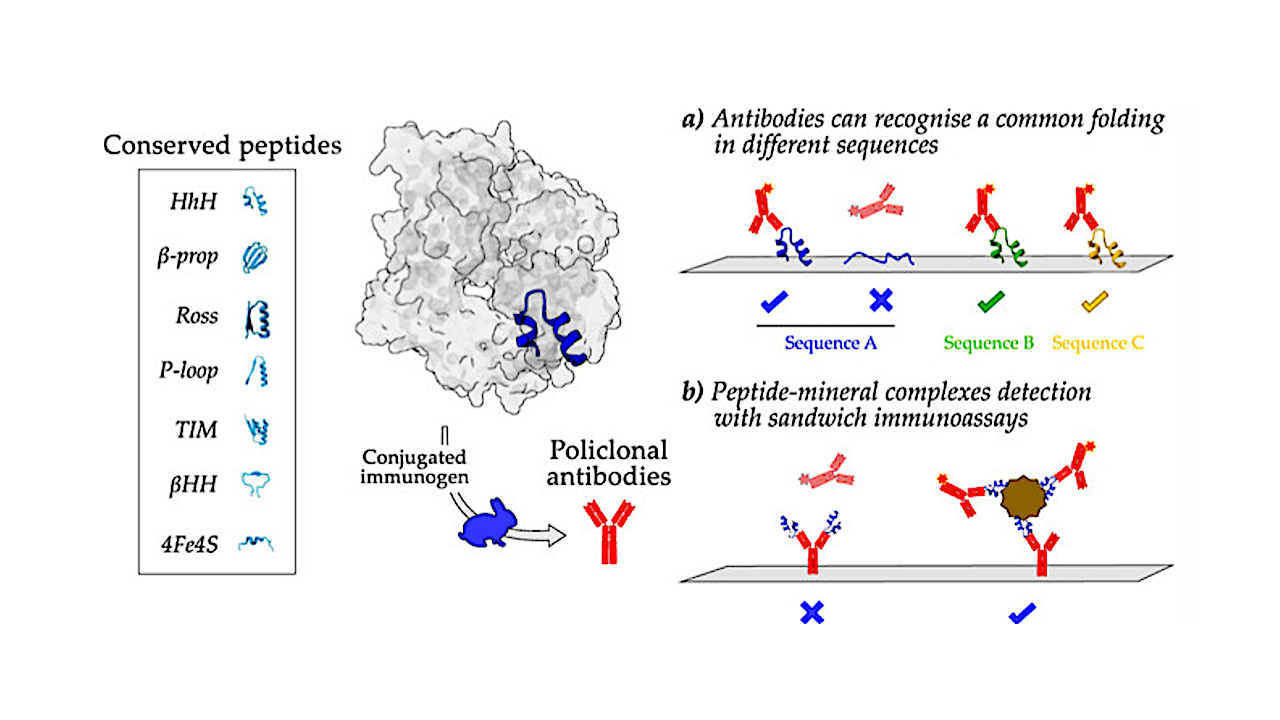
Their adsorption to minerals may contribute to their stabilization and preservation over time. To investigate the feasibility of conserved peptide sequences and structures as target biomarkers for the search for life on Mars or other planetary bodies, we conducted a bioinformatics selection of well-conserved ancient peptides and produced polyclonal antibodies for their detection using fluorescence microarray immunoassays.
Additionally, we explored how adsorbing peptides to Mars-representative minerals to form organomineral complexes could affect their immunological detection. The results demonstrated that the selected peptides exhibited autonomous folding, with some of them regaining their structure, even after denaturation.
Furthermore, their cognate antibodies detected their conformational features regardless of amino acid sequences, thereby broadening the spectrum of target peptide sequences.
While certain antibodies displayed unspecific binding to bare minerals, we validated that peptide–mineral complexes can be detected using sandwich immunoassays, as confirmed through desorption and competitive assays.
Consequently, we conclude that the diversity of peptide sequences and structures suitable for use as target biomarkers in astrobiology can be constrained to a few well conserved sets, and they can be detected even if they are adsorbed in organomineral complexes.

Folding prediction of the model peptides. (a) Peptide structure in the parental protein, indicated by their accession number (PDB) and location within the protein, showing ligands for HhH (single-stranded DNA), Ross (adenosine diphosphate), β-prop (glycerol), and 4Fe4S (4Fe4S cluster). (b) Most probable predicted structures of the model peptides in aqueous solution. — Anal Chem. via pubmed
Immunoanalytical Detection of Conserved Peptides: Refining the Universe of Biomarker Targets in Planetary Exploration, Anal Chem. 2024 Mar 26; 96(12): 4764–4773. Published online 2024 Mar 14. doi: 10.1021/acs.analchem.3c04165 (open access) via PubMed
Astrobiology