A Metabolic Secret Of Ethane-consuming Archaea Unraveled
Seeps on the deep seafloor naturally emit alkanes, which are pollutants that are potentially dangerous to life and act on global warming. Fortunately, the sediments around the seeps host microbes that act as a biological filter: They consume most of the alkanes before their release into the oceans and our atmosphere.
This so-called anaerobic oxidation of alkanes is an important yet poorly understood microbial process. Scientists from the Max Planck Institute for Marine Microbiology in Bremen, Germany, now present a study on the degradation of ethane, the second most abundant alkane in seeps. They characterized enzymes involved in the process and found that their reaction breaks an established dogma in the field of anaerobic biochemistry. Their results are published in Nature Communications.
A missing piece in the energy-retrieving machinery suspected from genomic data
The anaerobic oxidation of ethane was described a few years ago, and many of its secrets still have to be unraveled. “When drawing the chemical reactions of the pathway on paper, we found large gaps of uncharted biochemistry. We deduced that the involved organisms must acquire cellular energy through an unknown path”, explains first author Olivier Lemaire.
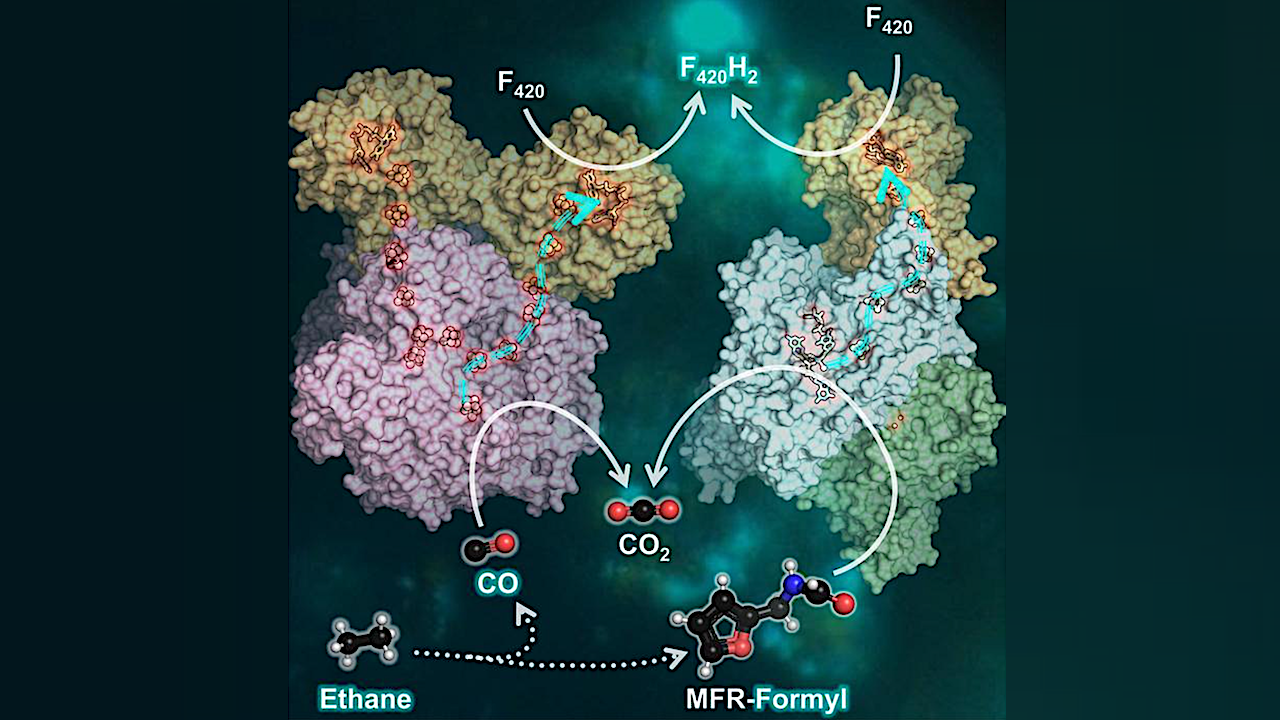
The two last enzymes of the process generate carbon dioxide (CO2) from the ethane. Other microbes use a protein called ferredoxin to take up electrons produced along the way. “That was also assumed in ethane oxidizers. However, when we looked at the genome of the microbes, we found that they don’t have the enzymatic tools to obtain cellular energy by the use of ferredoxin. Thus, something else must be at play.”
A challenging study achieved through a successful scientific collaboration
Solving this riddle was only possible thanks to a close collaboration within the Max Planck Institute for Marine Microbiology. Gunter Wegener and his team sampled the ethane-degrading microbial consortium from the deep sea and managed to culture it in the lab, despite this being a very demanding task.
Using these cultures, the group of Tristan Wagner managed to isolate and characterise the enzymes involved in ethane oxidation “Isolating enzymes from such a precious and complex microbial culture is a real challenge, but we managed with a lot of effort and meticulousness”, says Tristan Wagner.

a The pathway is based on studies described in Methanosarcinales7,8,30. The assembly of the ACDS (top) and Fwd (bottom) complexes are drawn in compliance with previous studies17,19,62. Arrows are coloured according to the corresponding metabolism, and dashed lines indicate multi-step transformations. White rounded rectangles framed by dotted lines illustrate internal channelling systems in which the substrates diffuse between both catalytic centres. Metallo-cofactor structures displayed in the inserts are derived from the deposited PDB models 1RU3 (acetyl-CoA synthase from Carboxydothermus hydrogenoformans63), 3CF4 (carbonylated ACDS α2ε2 subcomplex from M. barkeri17) and 5T5M (Fwd complex from M. wolfei19) with metals labelled in bold. cLys stands for carboxylysine. b Purification steps of the CODH component of the ACDS on native PAGE (left). (1) soluble extract; (2) anion exchange chromatography; (3, 4) hydrophobic exchange chromatography and (5) size-exclusion chromatography. The purified complex lacks the β, γ and δ subunits and, therefore, does not harbour the A-cluster and B12. c Purification steps of the Fwd/Fmd complex on native PAGE (left). (1) soluble extract; (2) anion exchange chromatography; (3) hydrophobic exchange chromatography and (4) size-exclusion chromatography. b, c An asterisk marks the band corresponding to the ECR11 on the native electrophoresis profile. b, c A denaturing (right) PAGE of the final enriched fractions. The electrophoresis profiles were similar for each purification. Source data are provided as a Source Data file.
A different enzymatic composition leads to a metabolic rewiring
The analyses now published show that both enzymes harbor an additional protein, electronically connected to the rest of the enzyme through a wire made of iron and sulfur. This subunit allows the use of an alternative electron acceptor: The F420, a molecule based on flavin, which is a class of chemicals also important to humans (as vitamin B2, for example).
“Such assemblies of CO2-forming enzymes and F420-reductases were never before described or suspected”, says Tristan Wagner. The researchers confirmed by additional experiments that both enzymes used F420 as an electron acceptor. “This discovery breaks a dogma in the scientific field of anaerobic metabolism, as it expands what these enzymes can do.”
“We suppose that the coupling of CO2-generation with F420 as electron acceptor might stimulate the entire process. The electrons are then transferred across the cell membrane to another microbe, reducing sulfate, which is a common principle in alkane-oxidizing consortia” says Gunter Wegener.
A milestone in the understanding of ethane degradation
By elucidating this metabolic riddle, Lemaire and his colleagues reveal a key aspect of the ethane-degrading microbes, which play an important role in the carbon cycle. It also shows that the knowledge gained from a few model organisms cannot be simply transposed to related species and that the enzymes involved can be more versatile than assumed.
“Our study illustrates how little we know about the metabolism of these microbes, which have lived on our planet for billions of years and can adapt to so many environments, and how important it is to understand them via experimental means”, Wagner concludes.
The study has a far-reaching impact as the alkane oxidation process performed by this type of microorganism is a crucial element of the biological filter existing in marine seeps, preventing massive effluxes of naturally produced alkanes in the atmosphere and seawater.
Ethane-oxidising archaea couple CO2 generation to F420 reduction, Nature Communications (open access)
Astrobiology