Genetic Encoding and Expression of RNA Origami Cytoskeletons in Synthetic Cells
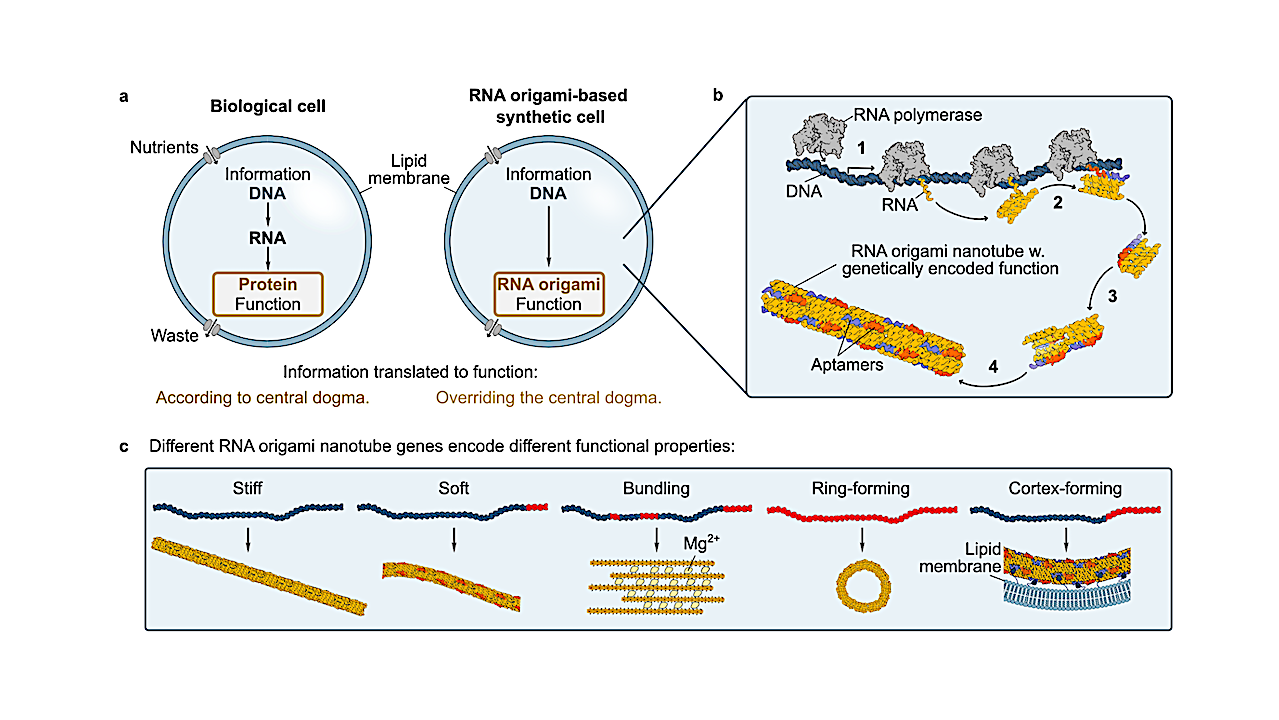
The central dogma at the core of molecular biology states that information flows from DNA to RNA and then to protein. Our research seeks to introduce a conceptually novel approach towards synthetic life by leveraging RNA origami, as an alternative to proteins, requiring only a single copying step between genetic information and function.
Here, we report the genetic encoding and expression of an RNA origami cytoskeleton-mimic within giant unilamellar lipid vesicles (GUVs). We design the first RNA origami tiles which fold co-transcriptionally from a DNA template and self-assemble into higher-order 3D RNA origami nanotubes at constant 37 ◦C in GUVs, where they reach several micrometers in length. Unlike pre-formed and encapsulated DNA cytoskeletons, these GUVs produce their own molecular hardware in an out-of-equilibrium process fuelled by nucleotide feeding.
To establish genotype-phenotype correlations, we investigate how sequence mutations govern the contour and persistence length of the RNA origami nanotubes with experiments and coarse-grained molecular-dynamics simulations, realizing a phenotypic transition to closed rings. Finally, we achieve RNA origami cortex formation and GUV deformation without chemical functionalization by introducing RNA aptamers into the tile design.
Altogether, this work pioneers the expression of RNA origami-based hardware in vesicles as a new approach towards active, evolvable and RNA-based synthetic cells.
Genetic encoding and expression of RNA origami cytoskeletons in synthetic cells, biorxiv.org
Astrobiology