Metabolism, Information, And Viability In A Simulated Physically-plausible Protocell
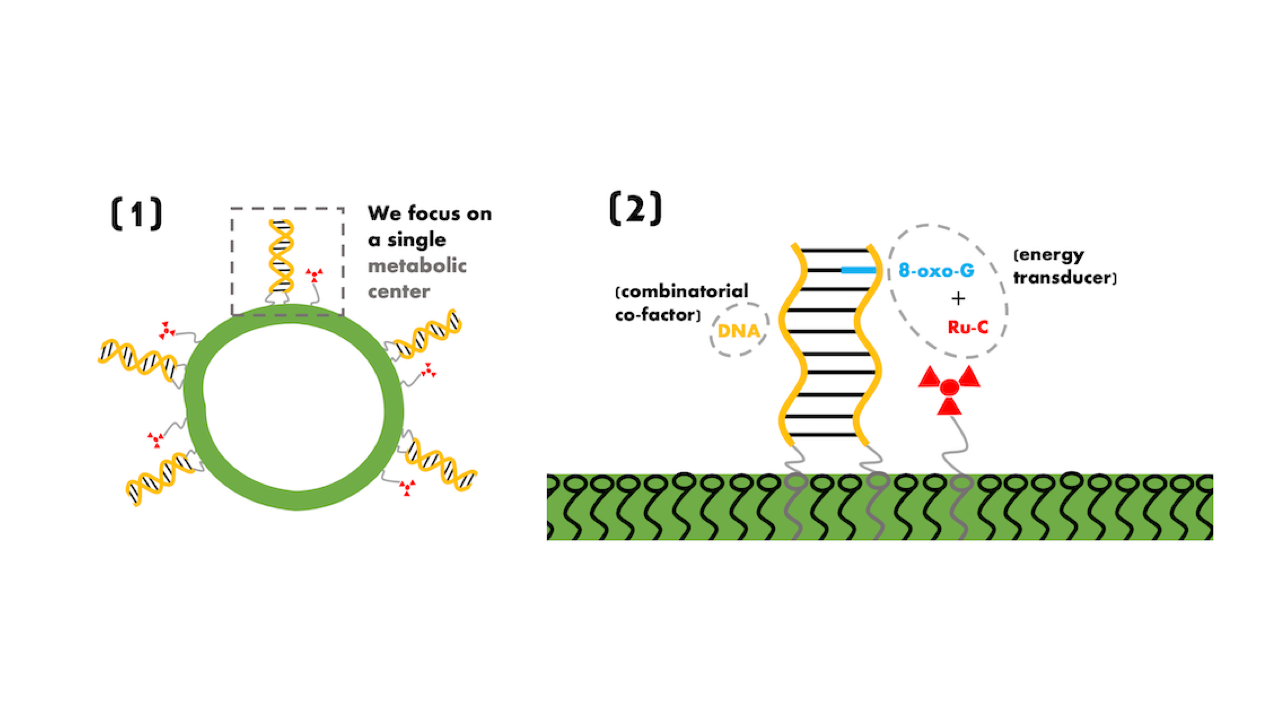
Critical experimental design issues connecting energy transduction and inheritable information within a protocell are explored and elucidated.
The protocell design utilizes a photo-driven energy transducer (a ruthenium complex) to turn resource molecules into building blocks, in a manner that is modulated by a combinatorial DNA-based co-factor. This co-factor molecule serves as part of an electron relay for the energy transduction mechanism, where the charge-transport rates depend on the sequence that contains an oxo-guanine. The co-factor also acts as a store of inheritable information due to its ability to replicate non-enzymatically through template-directed ligation.
Together, the energy transducer and the co-factor act as a metabolic catalyst that produces co-factor DNA building blocks as well as fatty acids (from picolinium ester and modified DNA oligomers), where the fatty acids self-assemble into vesicles on which exterior surface both the co-factor (DNA) and the energy transducer are anchored with hydrophobic tails.
Here we use simulations to study how the co-factor sequence determines its fitness as reflected by charge transfer and replication rates. To estimate the impact on the protocell, we compare these rates with previously measured metabolic rates from a similar system where the charge transfer is directly between the ruthenium complex and the oxo-guanine (without DNA replication and charge transport).
Replication and charge transport turn out to have different and often opposing sequence requirements. Functional information of the co-factor molecules is used to probe the feasibility of randomly picking co-factor sequences from a limited population of co-factors molecules, where a good co-factor can enhance both metabolic biomass production and its own replication rate.

Protocellular life-cycle: Panel (1) is a cartoon of the protocell with a fatty (decanoic) acid vesicle container decorated with anchored ruthenium complexes and DNA duplexes. Panel (2) shows droplet feeding of hydrophobic membrane precursors (picolineum esters). Panel (3) depicts absorption of the hydrophobic droplets into the membrane, where they are partly dissolved in the membrane, while the metabolism converts precursor lipids (picolinium ester) into lipids (decanoic acid). Panel (4) depicts ruthenium complex and the feeding of the DNA oligomer. Note that all precursors can be fed at once but are shown here as a two-step process for clarity. Panel (5) shows a completed (DNA) co-factor replication together with a completed fatty acid production, which results in membrane growth and vesicle destabilization. The original vesicle eventually breaks up and forms two new protocells, see Panel (6). — q-bio.MN
Kristoffer R. Thomsen, Artemy Kolchinsky, Steen Rasmussen
Subjects: Biological Physics (physics.bio-ph); Molecular Networks (q-bio.MN)
Cite as: arXiv:2405.04654 [physics.bio-ph] (or arXiv:2405.04654v1 [physics.bio-ph] for this version)
https://doi.org/10.48550/arXiv.2405.04654
Focus to learn more
Submission history
From: Steen Rasmussen
[v1] Tue, 7 May 2024 20:26:10 UTC (2,554 KB)
https://arxiv.org/abs/2405.04654
Astrobiology


![Discovery Of 1H-cyclopent[cd]indene (c-C11H8) In TMC-1 With The QUIJOTE Line Survey: A New Three-ringed Polycyclic Aromatic Hydrocarbon](https://astrobiology.com/wp-content/uploads/2026/01/Discovery-of-1H-cyclopentcdindene-1024x573.png)
