Convergence and Horizontal Gene Transfer Drive the Evolution of Anaerobic Methanotrophy in Archaea
Despite their large environmental impact and multiple independent emergences, the processes leading to the evolution of anaerobic methanotrophic archaea (ANME) remain unclear.
This work uses comparative metagenomics of a recently evolved but understudied ANME group, ‘Candidatus Methanovorans’ (ANME-3) to identify evolutionary processes and innovations at work in ANME which may be obscured in earlier evolved lineages. Within members of Methanovorans, we identified convergent evolution in carbon and energy metabolic genes as likely drivers of ANME evolution.
We also identified erosion of genes required for methylotrophic methanogenesis along with horizontal acquisition of multi-heme cytochromes and other loci uniquely associated with ANME. The assembly and comparative analysis of multiple Methanovorans genomes offers important functional context for understanding the niche-defining metabolic differences between methane-oxidizing ANME and their methanogen relatives.
Furthermore, this work illustrates the multiple evolutionary modes at play in the transition to a novel and globally important metabolic niche.

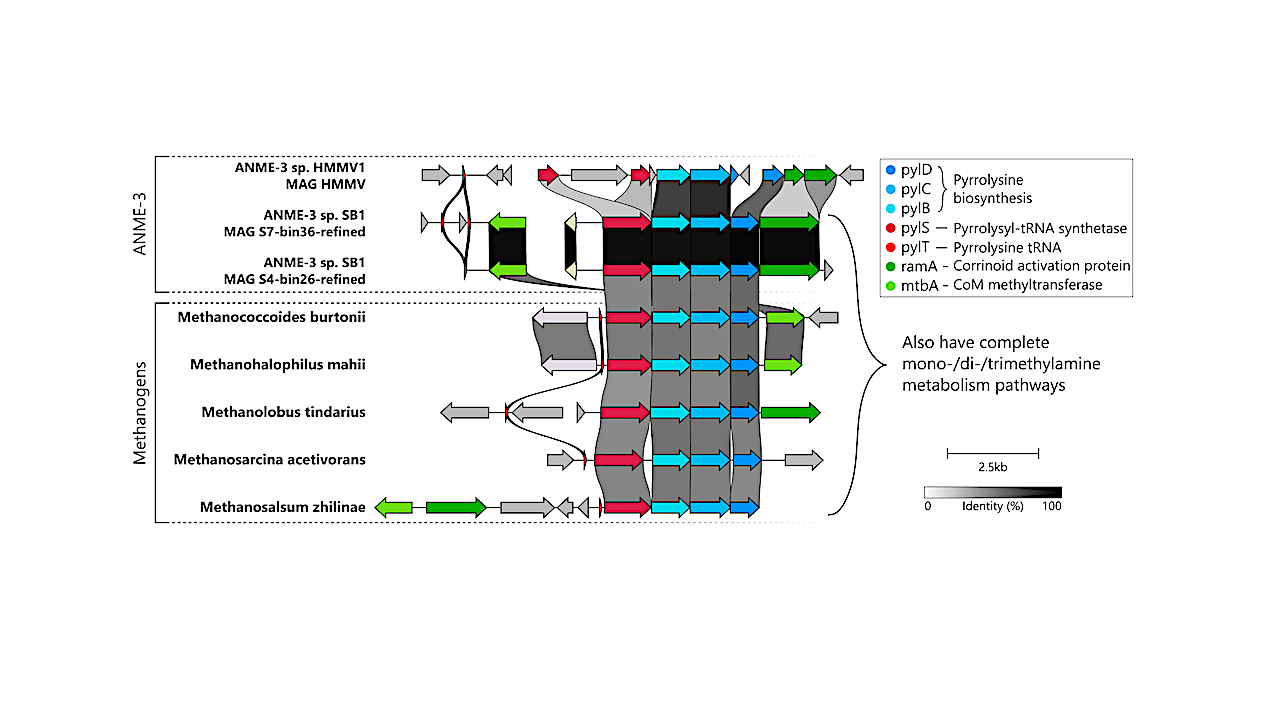
RNF complex organization (A), position in ANME metabolism (B), locus synteny (C), and gene phylogeny of rnfC (D) and rnfA (E). In panels A and C, subunits or genes colored purple concord with the estimated species phylogeny, subunits or genes colored green do not, and subunits or genes colored red are part of the locus only in ANME. The RNF complex (A) is seated within the membrane and couples electron transfer to sodium ion translocation. Gene organization of the rnf locus (C) is similar but not identical in ANME and methanogens. All studied genomes had the same organization of rnfCDGEAB and mmcA with the exception of Methanogasteraceae (ANME-2c). In addition, Methanocomedenaceae (ANME-2ab) and Methanovorans (ANME-3) loci also have two small transmembrane hairpin proteins and a b-type cytochrome, though the sequence similarity of these genes between Methanovorans and Methanocomedenaceae is low. Methanovorans and methanogenic Methanosarcinaceae have conserved flanking genes upstream of the locus, despite the extra genes in the Methanovorans loci. The gene phylogeny of rnfC (D) differs from the estimated species phylogeny, while the phylogeny of rnfA (E) matches the species phylogeny more closely. — biorxiv.org
Convergence and horizontal gene transfer drive the evolution of anaerobic methanotrophy in archaea, biorxiv.org
Astrobiology