Multi-substrate Specificity Shaped The Complex Evolution Of The Aminotransferase Family Across The Tree Of Life
Aminotransferases (ATs) are an ancient enzyme family that play central roles in core nitrogen metabolism essential to all organisms. However, many of the AT enzyme functions remain poorly defined, limiting our fundamental understanding of the nitrogen metabolic networks that exist in different organisms.
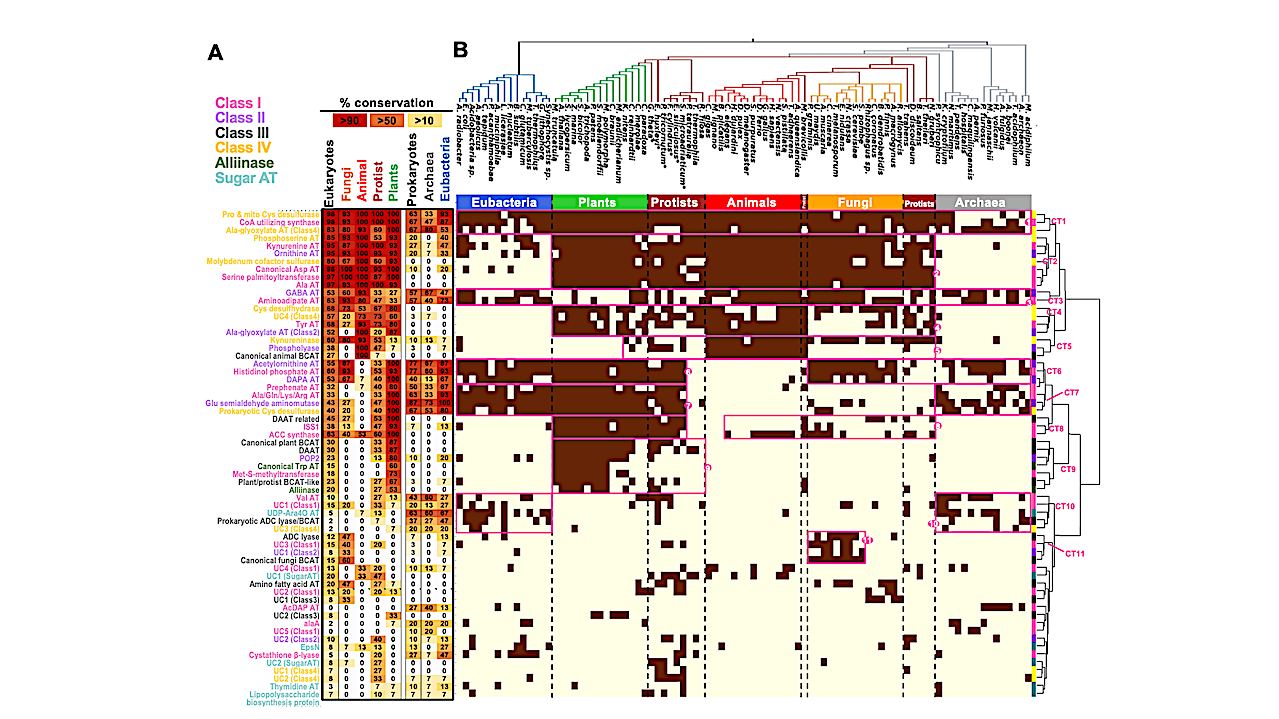
Here we traced the deep evolutionary history of the AT family by analyzing AT enzymes from 90 species spanning the tree of life (ToL). We found that each organism has maintained a relatively small and constant number of ATs. Mapping the distribution of ATs across the ToL uncovered that many essential AT reactions are carried out by taxon-specific AT enzymes due to wide-spread non-orthologous gene displacements.
This complex evolutionary history explains the difficulty of homology-based AT functional prediction. Biochemical characterizations of diverse aromatic ATs further revealed their broad substrate specificity, unlike other core metabolic enzymes that evolved to catalyze specific reactions today.
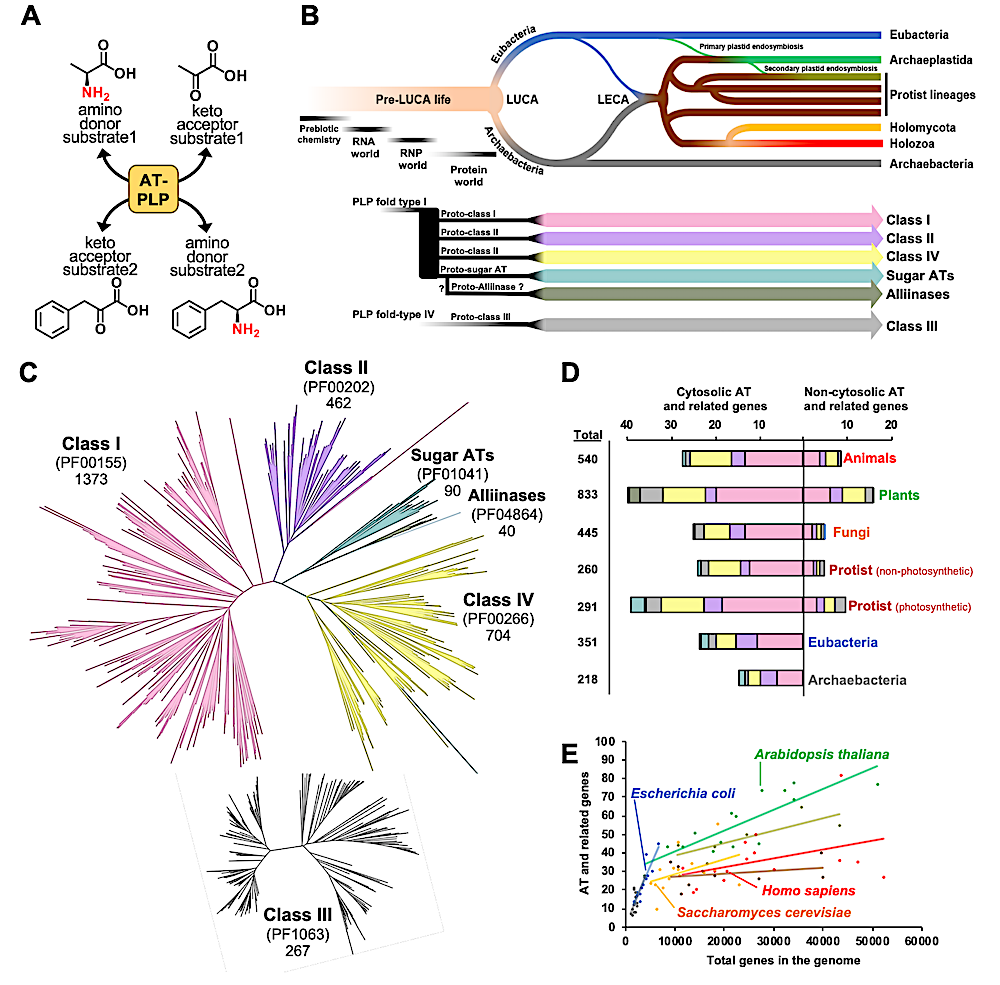
AT enzymes evolved with a limited copy number expansion across the Tree of Life (ToL). (A) Aminotransferases (ATs) are pyridoxal 5′-phosphate (PLP)-dependent enzymes, which catalyze reversible transamination reactions among at least four substrates. (B) Evolution of transamination since the origin of life to today. Pre-LUCA life likely had non-enzymatic, RNA, or RNP based transamination. At the interface of RNP and protein worlds, protein-based transaminases appeared in the form of proto-ATs classes. Proto-AT classes were inherited to LUCA and its descendants. (C) Phylogenetic analysis of AT candidate genes from seven Pfam domains known to contain ATs. Classes I, II and IV, alliinase and sugar ATs are part of PLP-fold type I, while class III is a part of the independently evolved PLP-fold type IV. (D) Average numbers, Pfam domain composition, and subcellular localization of AT and related genes from animals, plants, fungi, protists, eubacteria, and archaea. (E) The relationship between the number of AT and related genes versus total number of genes per species. Data corresponding to key model species is labeled and the lines show the overall trend per taxon. Blue, eubacteria; gray, archaea; brown, non-photosynthetic protists; army green, photosynthetic protists; orange, fungi; green, plants; red, animals. — biorxiv.org
Interestingly, however, we found that these AT enzymes that diverged over billion years share common signatures of multi-substrate specificity by employing different non-conserved active site residues. These findings illustrate that AT evolution had leveraged their inherent substrate promiscuity to maintain a small yet distinct set of multi-functional AT enzymes in different taxa.
This evolutionary history of versatile ATs likely contributed to the establishment of robust and diverse nitrogen metabolic networks that exist throughout the ToL. The study provides a critical foundation to systematically determine diverse AT functions and underlying nitrogen metabolic networks across the ToL.
Significance Statement The ToL-wide analyses of the ubiquitous aminotransferases (AT) family revealed that the broad substrate promiscuity of ATs, which is unusual for core metabolic enzymes, allowed recruitment of distinct, non-orthologous ATs to carry out essential AT reactions in different taxa but without increasing their copy numbers.
Some distantly related ATs were also found to exhibit a common signature of multi-substrate specificity by employing different non-conserved active site residues. The versatile evolutionary trajectory of the promiscuous AT enzyme family likely led to biochemical diversity of the robust nitrogen metabolic networks that exist among various extant organisms.
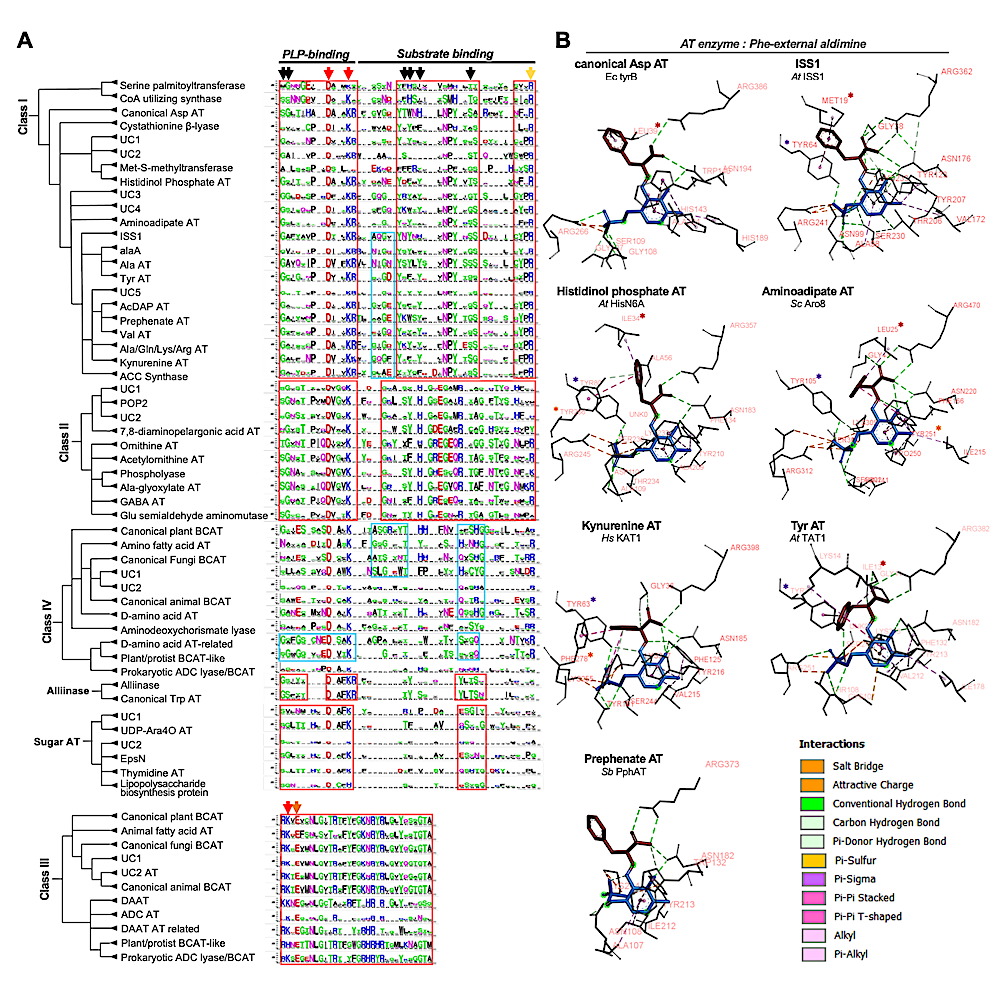
Distinct but functionally conserved residues underlie the conserved multi-substrate specificity among distantly related ATs. (A) Active site residues are extracted from representative enzymes from each AT group and are used to determine the consensus at those residue positions using the original multiple sequence alignment, which is shown in logo style132 1124 . Red and orange arrows show previously known well conserved residues for PLP-fold type I ATs, and black arrows show additional residues identified in this study. Red and blue boxes show motifs that are well conserved for an entire or a subset of each AT enzyme class, respectively. (B) Active site residues that interact with the PLP-Phe aldimine. The portions of the aldimine that corresponds to Phe and PLP are shown in red and blue sticks, respectively. Interacting residues are shown in black sticks and labeled with orange letters. The types of interactions (dashed lines) are colored based on the guide given on the figure. Residues having a conserved function in ligand interactions are marked with red, purple, and orange stars, respectively, as described in the main text. — biorxiv.org
doi: https://doi.org/10.1101/2024.03.19.585368
Multi-substrate specificity shaped the complex evolution of the aminotransferase family across the tree of life, biorxiv.org
Astrobiology, Genomics,