Genes Identified That Allow Bacteria To Thrive Despite Toxic Heavy Metal In Soil
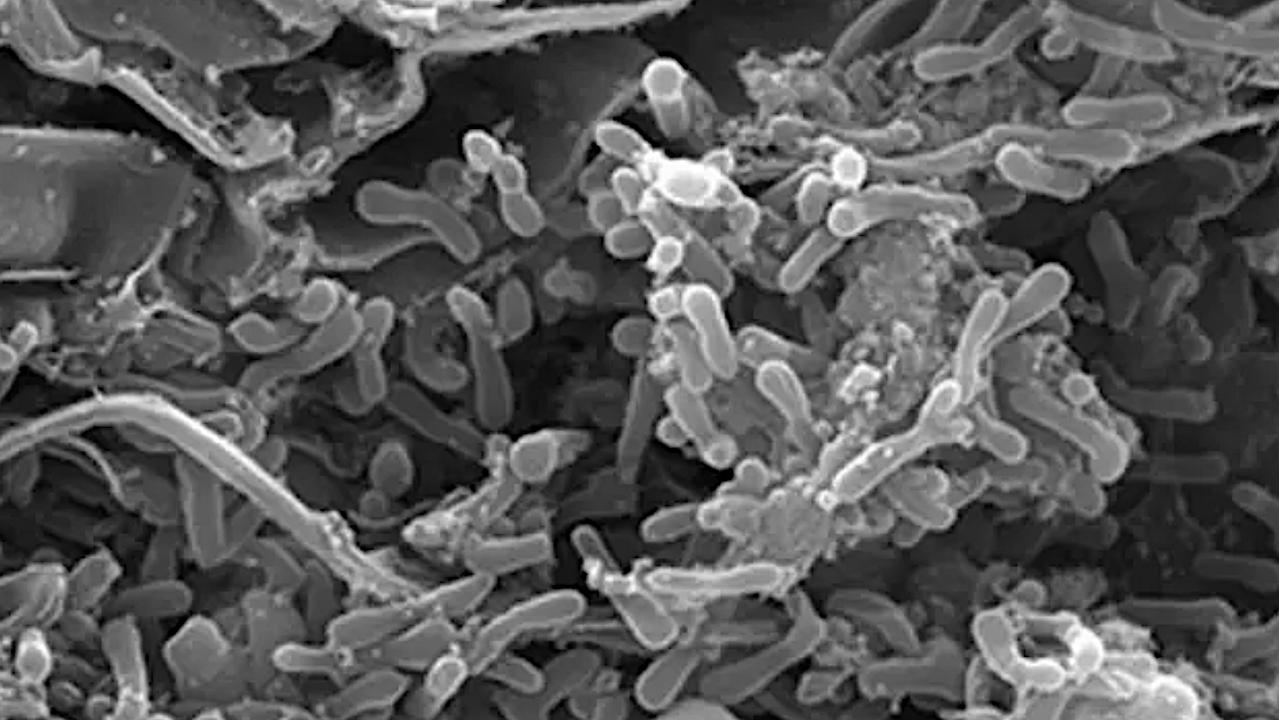
Some soil bacteria can acquire sets of genes that enable them to pump the heavy metal nickel out of their systems, a study has found. This enables the bacteria to not only thrive in otherwise toxic soils but help plants grow there as well.
A Washington State University-led research team pinpointed a set of genes in wild soil bacteria that allows them to do this in serpentine soils which have naturally high concentrations of toxic nickel. The genetic discovery, detailed in the journal Proceedings of the National Academies of Sciences, could help inform future bioremediation efforts that seek to return plants to polluted soils.
“We can say with certainty that these are the genes that are letting the bacteria survive the heavy metal exposure because if we take them away, they die. If we add them to a new bacterium that was sensitive to the heavy metal, all of the sudden it’s resistant,” said Stephanie Porter, the study’s senior author and a WSU evolutionary ecologist.
Soil bacteria called rhizobia are critical to legume plants, including commercial crops like soybean and alfalfa, since they symbiotically bond with roots and help the plants fix nitrogen, essentially fertilizing the plant.
For this study, Porter and her colleagues took samples of wild rhizobia bacteria from 55 grasslands in Oregon and California, some with nickel-heavy serpentine soils and some without. They conducted a range of genetic analysis and found a set of genes, called the nickel resistance operon, were necessary to allow the bacteria to survive exposure to the heavy metal.
They also found that the adaptation was finely tuned to the level of nickel in the soil. Bacteria from areas with high nickel concentrations had versions of the genes that conferred more tolerance, while those from areas with lower amounts had genes that were not as effective for tolerating higher levels of nickel.
“It’s like there’s this very beautiful matching between these rhizobia and their habitats,” Porter said. “It’s an exquisite evolutionary story about how diversity arises and is maintained in nature—to very closely match the level of challenge that these organisms face.”
The team is investigating further the way the bacteria achieve this adaptation through what is known as “horizontal gene transfer.” Unlike animals, bacteria do not only transfer genetic information from parent to child. They can also share “mobile” sets of genes with peer bacteria just by coming in close contact with them.
Porter likens this process to downloading an app on a smartphone, where one bacterium cell joins up with another in the environment, and they exchange packets of information, essentially sets of genes. The bacterium then “downloads” the information and the new DNA becomes part of that organism’s genome.
Many kinds of bacteria do this to adapt to different environments, said co-author Angeliqua Montoya, a WSU Ph.D. candidate in Porter’s lab. This includes some bacteria which are problematic for humans, such as the harmful bacteria that can acquire resistance to antibiotics.
“There is a whole spectrum of traits that these mobile elements confer in bacteria,” Montoya said.
The researchers are betting that by better understanding these mobile genetic elements, some of these traits can be harnessed to use microbes to help overcome challenges, like polluted soils, that are having increasing impacts.
The work received support from the National Science Foundation, the Murdock Charitable Trust, and Washington State University. Other researchers on the study include first author Hanna Kehlet Delgado and co-authors Camille Wendlandt, Chrisopher Dexheimer, Miles Roberts and Maren Friesen from WSU; Kyson Jenson and Joel Griffitts from Bringham Young University; and Lorena Torres Martinez from St. Mary’s College of Maryland.
The evolutionary genomics of adaptation to stress in wild rhizobium bacteria, PNAS
Astrobiology