Extremely Acidic Proteomes And Metabolic Flexibility In Bacteria And Highly Diversified Archaea Thriving In Geothermal Chaotropic Brines
Few described archaeal, and fewer bacterial, lineages thrive at salt-saturating conditions, such as solar saltern crystallizers (salinity above 30%-w/v).
They accumulate molar K+ cytoplasmic concentrations to maintain osmotic balance (‘salt-in’ strategy), and have proteins adaptively enriched in negatively charged, acidic amino acids. Here, we analyzed metagenomes and metagenome-assembled genomes (MAGs) from geothermally influenced hypersaline ecosystems with increasing chaotropicity in the Danakil Depression.
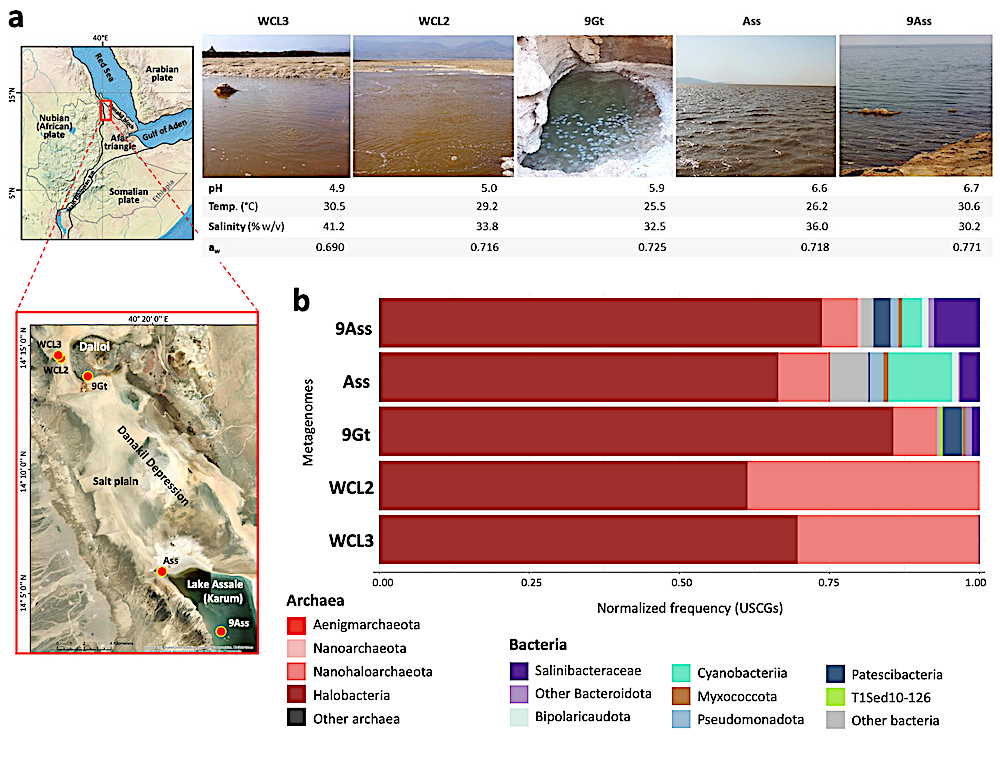
Microbial community composition inferred from metagenomes of polyextreme chaotropic ecosystems in the north Danakil Depression. a, Sampling sites around the Dallol proto-volcano and Lake Assale or Karum in the north Danakil Depression, Ethiopia; some major abiotic parameters for each system are shown (aw, water activity). b, Global microbial community composition at high-rank taxonomic level of sampled polyextreme ecosystems inferred from the normalized frequency of universal single-copy genes (USCGs; selection of ribosomal proteins expressed in RPKM). Note that DAL-WCL2 and WCL3 are composed of 99% archaea (Halobacteriota and Nanohaloarchaeota; classification according to GTDB r214). — biorxiv.org
Normalized abundances of universal single-copy genes confirmed that haloarchaea and Nanohaloarchaeota encompass 99% of microbial communities in the near life-limiting conditions of the Western-Canyon Lakes (WCLs). Danakil metagenome- and MAG-inferred proteomes, compared to those of freshwater, seawater and solar saltern ponds up to saturation (6-14-32% salinity), showed that WCL archaea encode the most acidic proteomes ever observed (median protein isoelectric points ≤4.4).
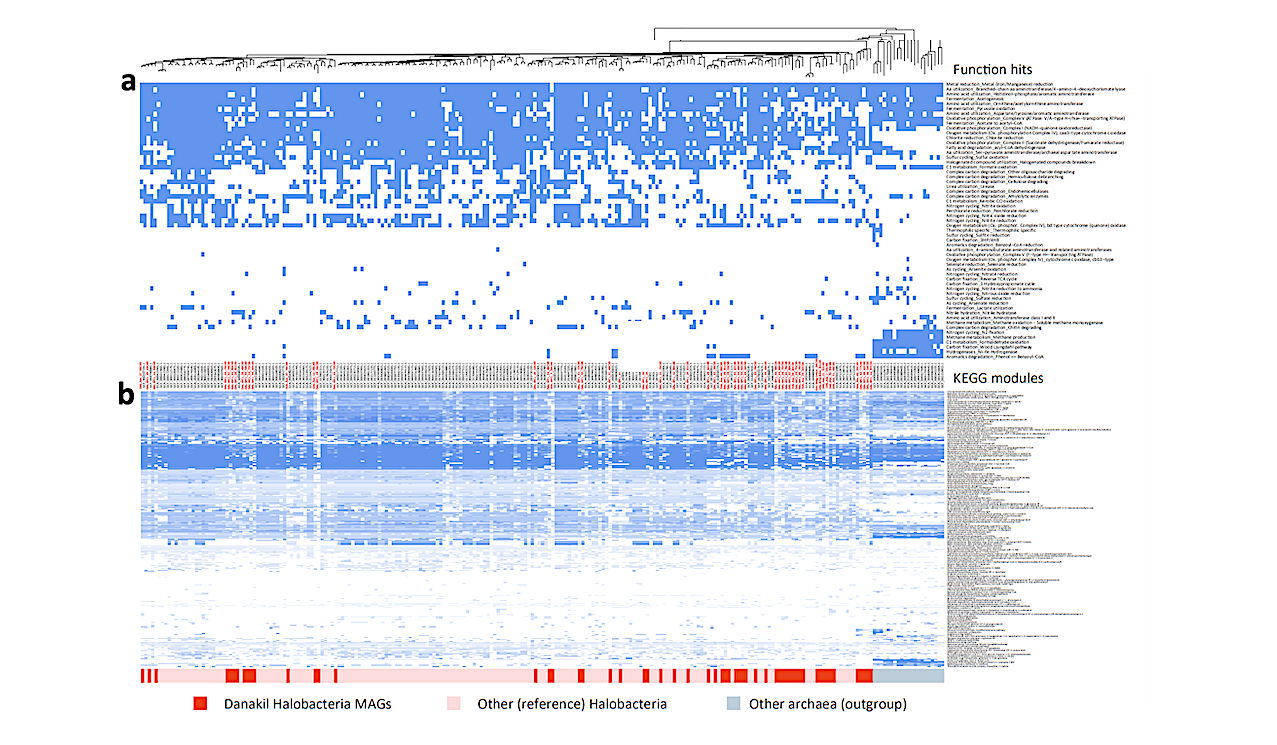
We identified previously undescribed Halobacteria families as well as an Aenigmatarchaeota family and a bacterial phylum independently adapted to extreme halophily. Despite phylum-level diversity decreasing with increasing salinity-chaotropicity, and unlike in solar salterns, adapted archaea exceedingly diversified in Danakil ecosystems, challenging the notion of decreasing diversity under extreme conditions.
Metabolic flexibility to utilize multiple energy and carbon resources generated by local hydrothermalism along feast-and-famine strategies seemingly shape microbial diversity in these ecosystems near life limits.
doi: https://doi.org/10.1101/2024.03.10.584303
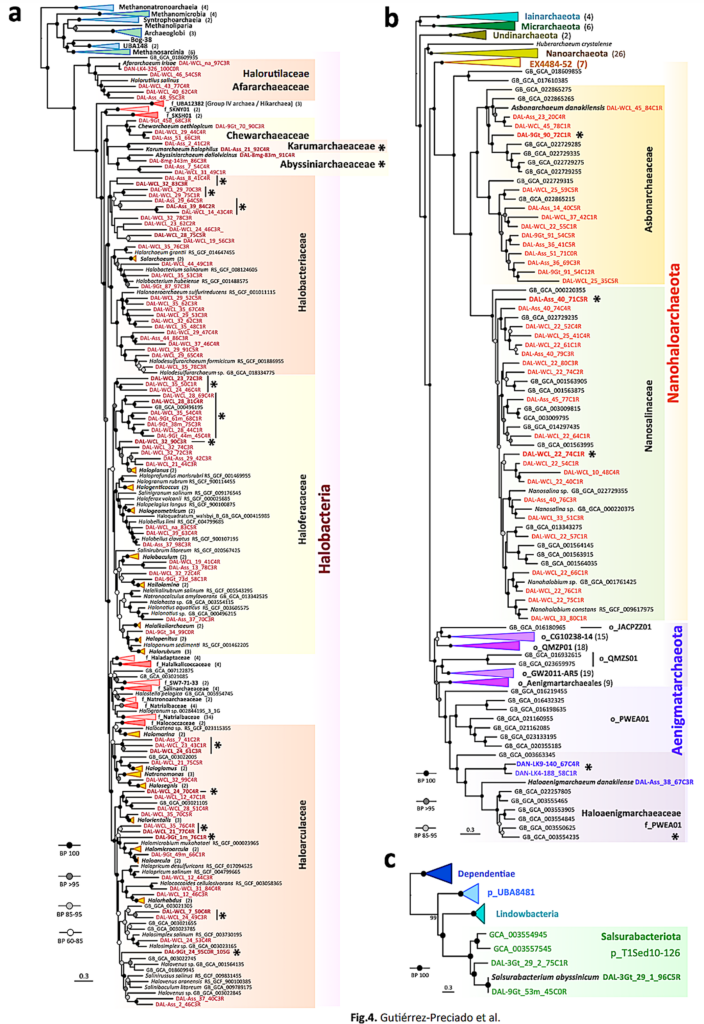
Phylogenomic trees of newly identified archaeal and bacterial MAGs in hypersaline chaotropic north Danakil ecosystems. a, Phylogenomic tree of archaeal MAGs affiliating to the class Halobacteria (Halobacteriota). b, Phylogenomic tree of MAGs affiliating to the Nanohaloarchaeota and Aenigmatarchaeota (DPANN supergroup). c, Phylogenomic analysis of the bacterial phylum Candidatus Salsurabacteriota (p_T1Sed10-126), grouping seemingly extreme halophilic members. Bootstrap value ranges are indicated at nodes. Names of MAGs assembled from our Danakil metagenomes are highlighted in color; reference sequences are indicated in black. Some taxa were collapsed to facilitate visualization of trees (detailed trees are shown in Supplementary Figs. 4-6); the number of total representatives included in collapsed taxa is given in brackets. Asterisks indicate genera and families newly identified in this study. — biorxiv.org
Astrobiology