Tardigrade Genomes Reveal The Secrets Of Extreme Survival
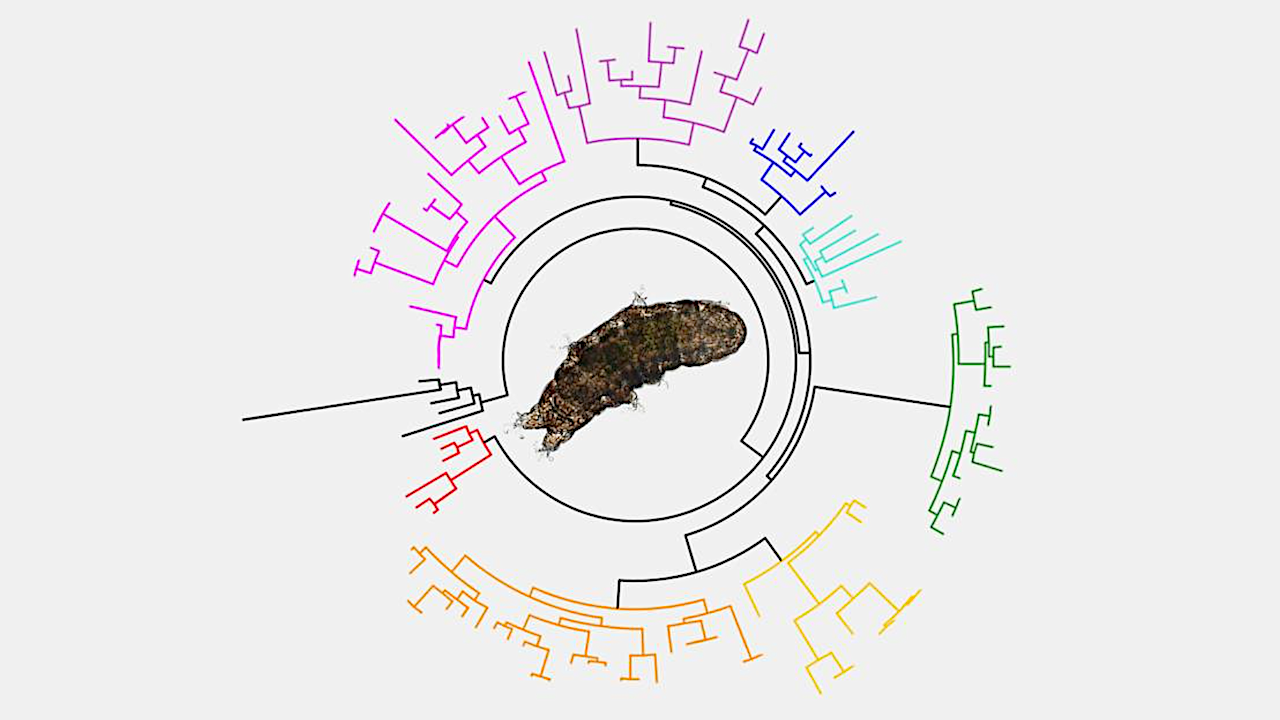
Tardigrades may be nature’s ultimate survivors. While these tiny, nearly translucent animals are easily overlooked, they represent a diverse group that has successfully colonized freshwater, marine, and terrestrial environments on every continent, including Antarctica.
Commonly known as “water bears”, these unusual creatures may be among the most resilient organisms on the planet thanks to their unparalleled ability to survive extreme conditions, with various species being resistant to drought, high doses of radiation, low oxygen environments, and both high and low temperatures and pressures.
nsive understanding of the origins and history of these unique adaptations has remained elusive. In a new study published in Genome Biology and Evolution, scientists at Keio University Institute for Advanced Biosciences, the University of Oslo Natural History Museum, and the University of Bristol reveal a surprisingly intricate network of gene duplications and losses associated with tardigrade extremotolerance, highlighting the complex genetic landscape that drives modern tardigrade ecology.
As one form of extremotolerance, tardigrades can survive almost complete desiccation by entering a dormant state called anhydrobiosis (i.e., life without water), which allows them to reversibly halt their metabolism. Multiple tardigrade-specific gene families were previously found to be associated with anhydrobiosis. Three of these gene families are referred to as cytosolic, mitochondrial, and secretory abundant heat soluble proteins (CAHS, MAHS, and SAHS, respectively) based on the cellular location in which the proteins are expressed.
Some tardigrades appear to possess a variant pathway that involves two families of abundant heat soluble proteins first identified in the tardigrade Echiniscus testudo and usually referred to as EtAHS alpha and beta. Tardigrades also possess stress resistance genes that can be found in animals more broadly, such as the meiotic recombination 11 (MRE11) gene, which has been implicated in desiccation tolerance in other animals. Unfortunately, since the identification of these gene families, limited information has been available from most tardigrade lineages, making it difficult to draw conclusions on their origins, history, and ecological implications.
To better shed light on the evolution of tardigrade extremotolerance, the authors of the new study—James Fleming, Davide Pisani and Kazuharu Arakawa—identified sequences from these six gene families across 13 tardigrade genera, including representatives from both of the major tardigrade lineages, the Eutardigrades and Heterotardigrades. Their analysis revealed 74 CAHS, 8 MAHS, 29 SAHS, 22 EtAHS alpha, 18 EtAHS beta, and 21 MRE11 sequences, allowing them to build the first tardigrade phylogenies for these gene families.
As resistance to desiccation is likely to have emerged as an adaptation to terrestrial environments, the authors assumed that they would find a link between gene duplications and losses in these gene families and habitat changes within tardigrades. “When we began the work, we expected to find that each clade would be clearly grouped around ancient duplications, with few independent losses. That would help us easily tie them to an understanding of modern habitats and ecology,” says the study’s lead author, James Fleming. “It’s an intuitive hypothesis,” he continues, “that the evolution of the duplications of these desiccation-related genes should, in theory, contain remnants of the ecological history of these organisms, although, in reality, this turned out to be overly simplistic.”
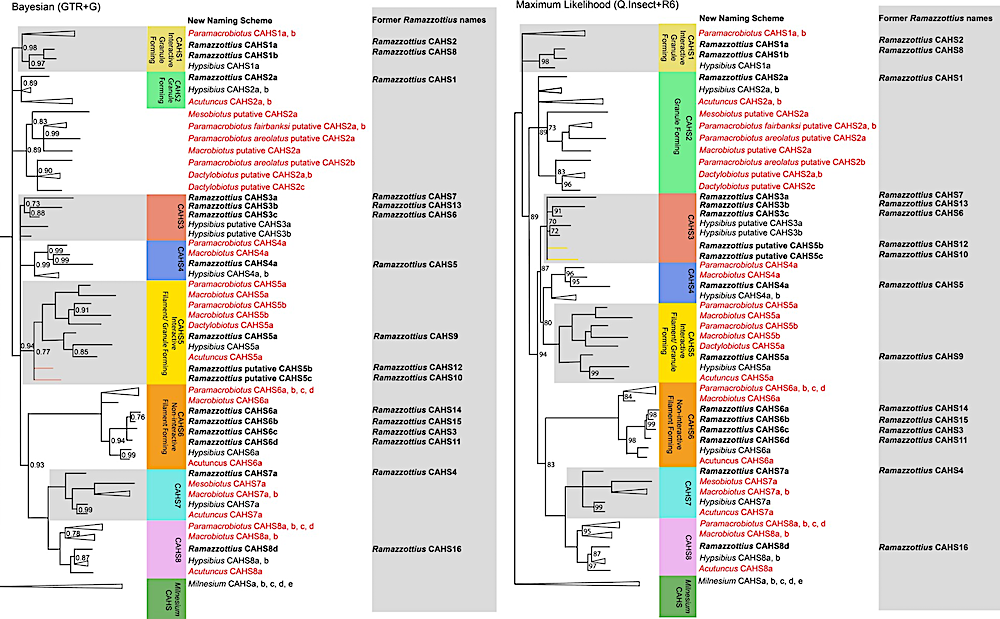
Bayesian consensus and maximum likelihood phylogenies depicting the CAHS protein family within Tardigrada. The former names of the Ramazzottius CAHS proteins are depicted to the right of each Ramazzottius CAHS protein, alongside the phylogenetically informative nomenclature for the CAHS protein introduced within this paper. The sequences introduced in this study are colored in red, and the sequences recovered from prior studies are in black. PPs <1 are marked on the Bayesian phylogeny, and PPs <0.7 are reduced to polytomies. UBS values <100 are marked on the maximum likelihood phylogeny, and nodes with <70 support are reduced to polytomies. The nodes comprising only independent duplications have been collapsed for readability, unless these independent duplications belong to Ramazzottius, where they have been retained for clarity in nomenclature revision. The conflicts between the Bayesian and the maximum likelihood phylogeny have been highlighted by coloring the branches of the corresponding group in the opposite phylogeny. The assumed ancestral functional annotations have been added to the group where sequences have been functionally characterized, but individual variation is also present within groups: Hypsibius CAHS2b, for example, forms fibrous gels but is a member of the granule-forming group. — SMBE Journals
Instead, the authors were surprised by the sheer number of independent duplications of heat-soluble genes, which painted a much more complex picture of anhydrobiosis-related gene evolution. Notably, however, there was no clear link between strongly anhydrobiotic species and the number of anhydrobiosis-related genes a species possessed. “What we found was far more exciting,” says Fleming, “a complex network of independent gains and losses that does not necessarily correlate to modern terrestrial species ecologies.”
Despite the lack of a relationship between gene duplications and tardigrade ecology, the study did provide crucial insight into the major transitions that led to the acquisition of anhydrobiosis. The distinct distributions of gene families across the two major groups of tardigrades—CAHS, MAHS, and SAHS in the Eutardigrades and EtAHS alpha and beta in the Heterotardigrades—suggest that two independent transitions from marine to limno-terrestrial environments occurred within tardigrades, once in the Eutardigrade ancestor and once within the Heterotardigrades.
This research marks a significant step forward in our understanding of the evolution of anhydrobiosis in tardigrades. It also provides a foundation for future studies into tardigrade extremotolerance, which will require the continued development of genomic resources from more diverse tardigrade lineages. “We unfortunately have no representatives from several important families, such as the Isohypsibiidae, and this does limit how firmly we can stand by our conclusions,” notes Fleming. “With more freshwater and marine tardigrade samples, we will be better able to appreciate the adaptations of terrestrial members of the group.”
Unfortunately, some tardigrades can be especially elusive, presenting a major obstacle to such studies. As an example, Tanarctus bubulubus, one of Fleming’s favorite tardigrades, is too small to see with the naked eye and is found only in sediment in the North Atlantic at depths of around 150 m. “Hopefully,” says Fleming, “large-scale sequencing initiatives through the Earth Biogenome Project will steadily bridge this gap in our understanding, and it’s an effort I’m excited to see continue.”
The Evolution of Temperature and Desiccation-Related Protein Families in Tardigrada Reveals a Complex Acquisition of Extremotolerance, Genome Biology and Evolution(open access)
Astrobiology, genomics,