A High-quality Chromosomal Genome Assembly Of The Sea Cucumber Chiridota Heheva And Its Hydrothermal Adaptation

Background – Chiridota heheva is a cosmopolitan holothurian well adapted to diverse deep-sea ecosystems, especially chemosynthetic environments. Besides high hydrostatic pressure and limited light, high concentrations of metal ions also represent harsh conditions in hydrothermal environments. Few holothurian species can live in such extreme conditions. Therefore, it is valuable to elucidate the adaptive genetic mechanisms of C. heheva in hydrothermal environments.
Findings – Herein, we report a high-quality reference genome assembly of C. heheva from the Kairei vent, which is the first chromosome-level genome of Apodida. The chromosome-level genome size was 1.43 Gb, with a scaffold N50 of 53.24 Mb and BUSCO completeness score of 94.5%. Contig sequences were clustered, ordered, and assembled into 19 natural chromosomes. Comparative genome analysis found that the expanded gene families and positively selected genes of C. heheva were involved in the DNA damage repair process. The expanded gene families and the unique genes contributed to maintaining iron homeostasis in an iron-enriched environment. The positively selected gene RFC2 with 10 positively selected sites played an essential role in DNA repair under extreme environments.
Conclusions – This first chromosome-level genome assembly of C. heheva reveals the hydrothermal adaptation of holothurians. As the first chromosome-level genome of order Apodida, this genome will provide the resource for investigating the evolution of class Holothuroidea.
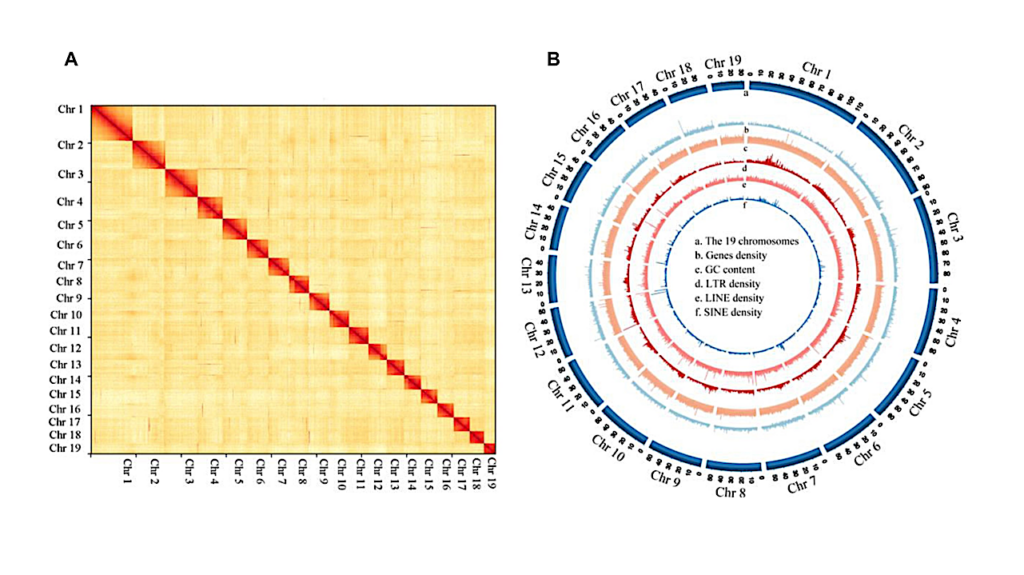
Genome assembly and sequencing analysis of the Kairei vent Chiridota heheva. (A) Hi-C interaction heatmap. (B) High-quality assembly of 19 chromosomes with genes coverage, GC content, and repetitive elements of long terminal repeat, LINE, and SINE. – Gigascience
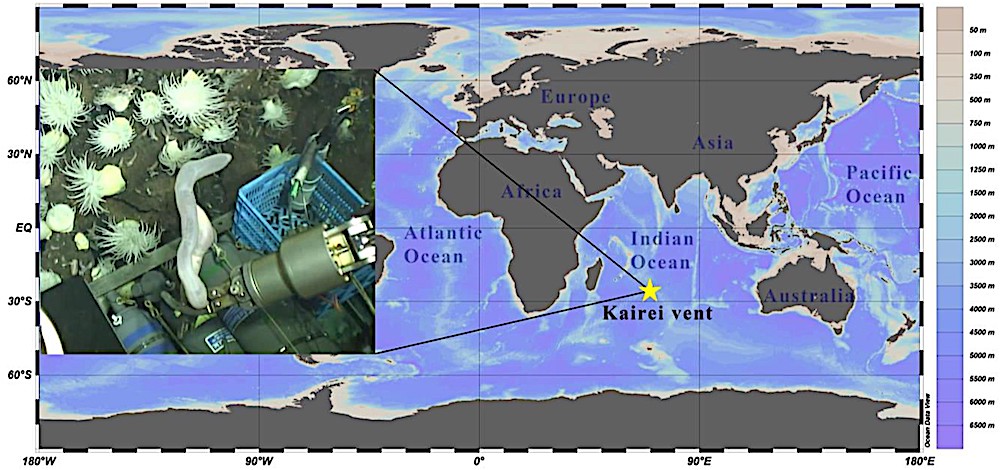
The sampling site at the Kairei vent field of the Indian Ocean and the photo in situ at a depth of 2,428 m. Credit: IDSSE Deep-sea Scientific Research Image and Video Database.
A high-quality chromosomal genome assembly of the sea cucumber Chiridota heheva and its hydrothermal adaptation (open access)
Yujin Pu, Yang Zhou, Jun Liu, Haibin Zhang
GigaScience, Volume 13, 2024, giad107, https://doi.org/10.1093/gigascience/giad107
Published: 04 January 2024 Article history
Astrobiology