Planetary Scale Assay Of Earth Life: The Monarch Initiative In 2024: An Analytic Platform Integrating Phenotypes, Genes And Diseases Across Species

Editor’s note: As we make plans to do orbital and surface sorties on other worlds searching for life it is certainly useful to use Earth and its diverse biota as an analog to figure out how to do this. If we do find life on a world we’ll want to catalog what we find based on habitat, physiology, and genomics. Using planet Earth as a testbed or analog for planetary-level genomic modeling is a good way to learn how to do the same thing on other worlds – and we can start doing it right now.
Bridging the gap between genetic variations, environmental determinants, and phenotypic outcomes is critical for supporting clinical diagnosis and understanding mechanisms of diseases. It requires integrating open data at a global scale.
The Monarch Initiative advances these goals by developing open ontologies, semantic data models, and knowledge graphs for translational research. The Monarch App is an integrated platform combining data about genes, phenotypes, and diseases across species.
Monarch’s APIs enable access to carefully curated datasets and advanced analysis tools that support the understanding and diagnosis of disease for diverse applications such as variant prioritization, deep phenotyping, and patient profile-matching. We have migrated our system into a scalable, cloud-based infrastructure; simplified Monarch’s data ingestion and knowledge graph integration systems; enhanced data mapping and integration standards; and developed a new user interface with novel search and graph navigation features.
Furthermore, we advanced Monarch’s analytic tools by developing a customized plugin for OpenAI’s ChatGPT to increase the reliability of its responses about phenotypic data, allowing us to interrogate the knowledge in the Monarch graph using state-of-the-art Large Language Models. The resources of the Monarch Initiative can be found at monarchinitiative.org and its corresponding code repository at github.com/monarch-initiative/monarch-app.
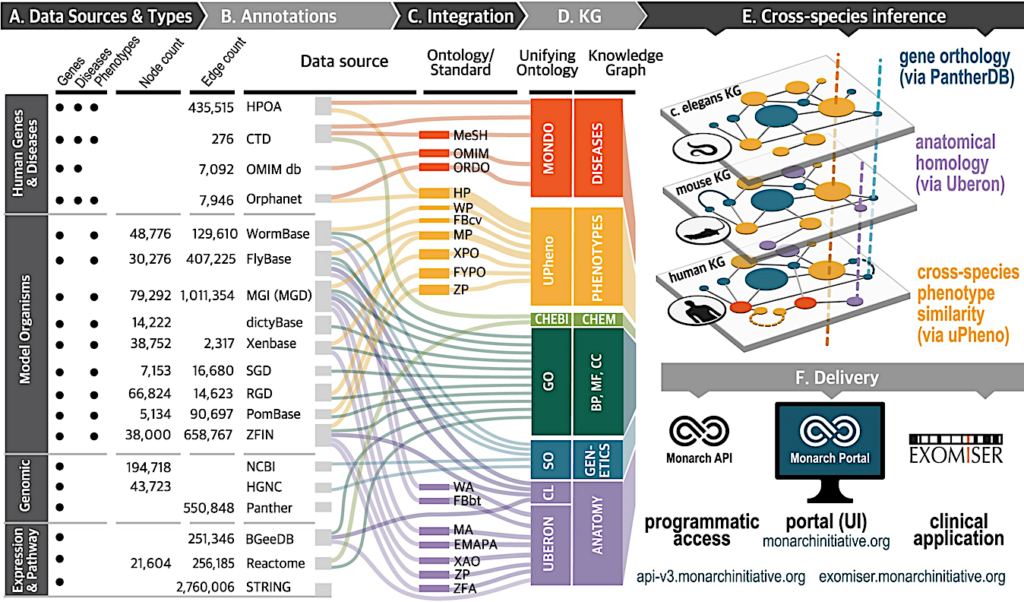
Data harmonization within the Monarch KG. The three primary data types in the Monarch KG are genes, diseases and phenotypes (A).This image details their entity (node) and link (edge) counts and the unifying ontologies (D) by which the source data (B) and ontologies (C) are harmonized. Cross-species inference (E) is accomplished via gene orthology, homology and phenotype similarity. Content dissemination (F) is via API, the Monarch UI and within the clinical application Exomiser. Note that the figure expresses only a portion of the integrated ontologies (column C). For a comprehensive list see PHENIO documentation (linked below). In Column D, GO: Gene Ontology; BP: Biological Process; MF: Molecular Function; CC: Cellular Component.
The Monarch Initiative in 2024: an analytic platform integrating phenotypes, genes and diseases across species, Nucleic Acids Research (open access)
Astrobiology, genomics,