Modeling Genome-scale Knowledge In Earth’s Global Ocean
Editor’s note: As we make plans to do orbital and surface sorties on other worlds searching for life it is certainly useful to use Earth and its diverse biota as an analog to figure out how to do this. If we do find life on a world we’ll want to catalog what we find based on habitat, physiology, and genomics. Using planet Earth as a testbed for planetary-level genomic modeling is a good way to learn how to do the same thing on other worlds – and we can start doing it right now.
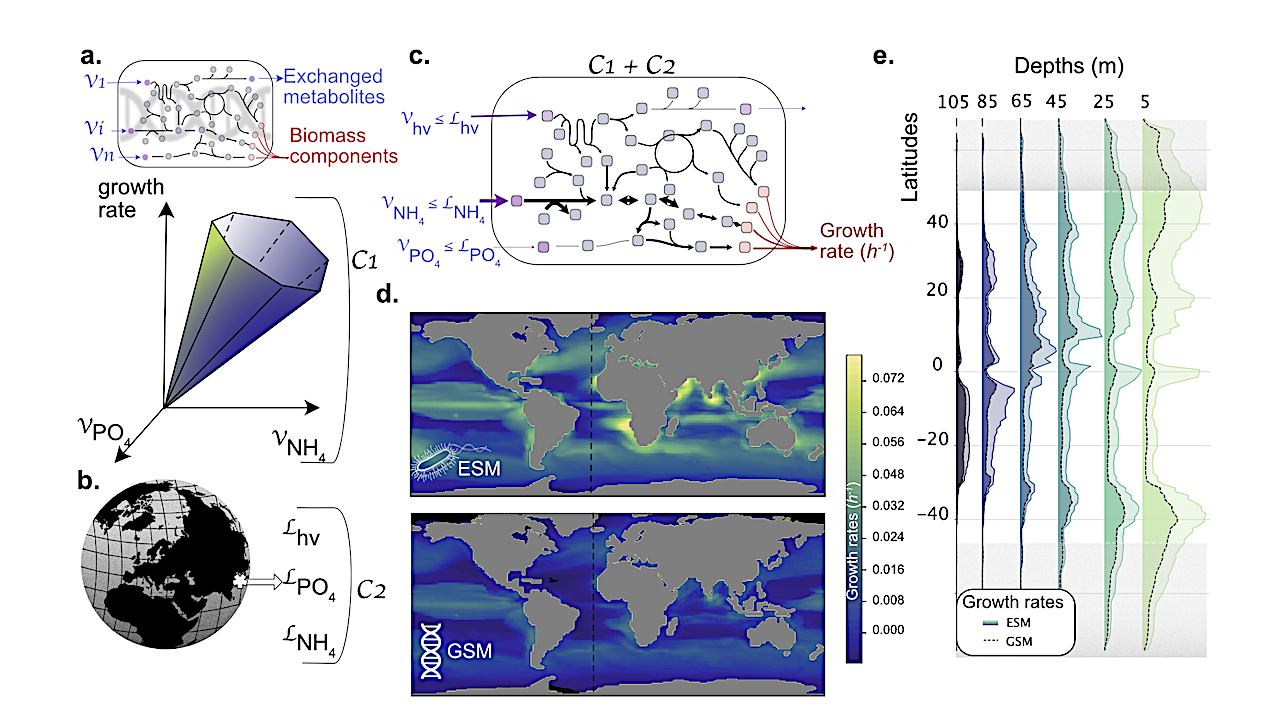
Earth System Models (ESMs) highly simplify their representation of biological processes, leading to major uncertainty in climate change impacts.
Despite a growing understanding of molecular networks from genomic data, describing how changing phytoplankton physiology affects the production of key metabolites remains elusive.
Here we embed a genome-scale model within a state-of-the-art ESM to deliver an integrated understanding of how gradients of nutritional constraints modulate metabolic reactions and molecular physiology.
Applied to the prevalent marine cyanobacteria Prochlorococcus, we find that glycogen and lipid storage can be understood as a consequence of acclimation to environmental gradients.
Given the pressing need to assess how biological diversity influences biogeochemical functions, genome-enabled ESMs allow the quantification of the contribution of modeled organisms to the production of dissolved organic carbon and its molecular composition.
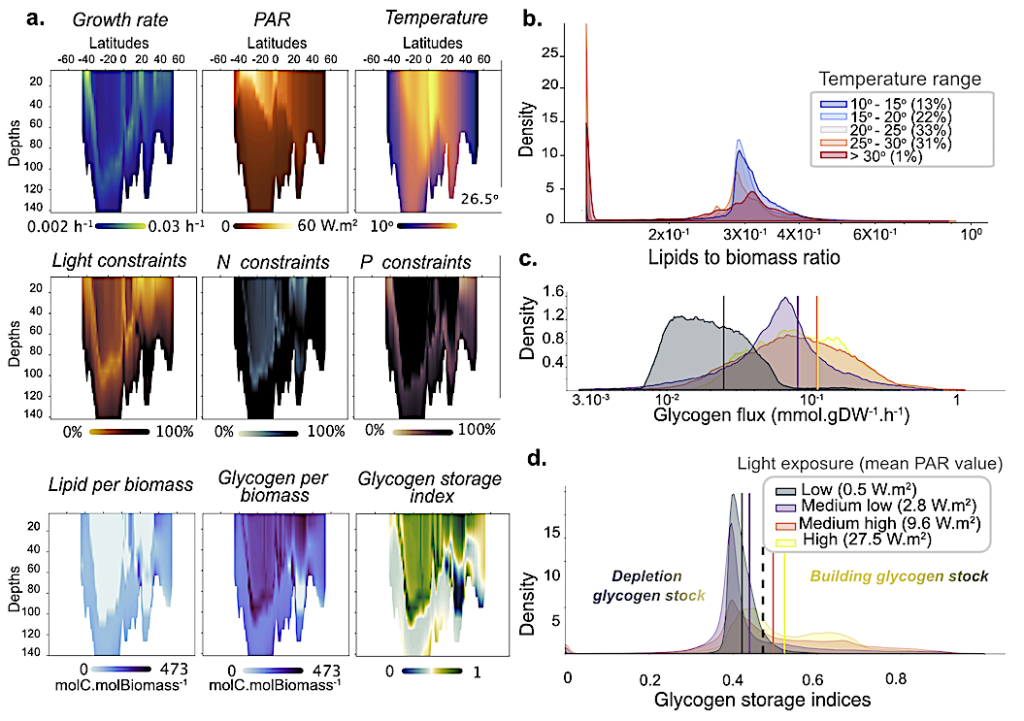
Investigation of Prochlorococcus MED4 genome-scale model fitness and acclimation strategies across the global ocean (more than 106 estimations). a |Description of Prochlorococcus MED4 genome-scale behavior across the Atlantic Ocean transect (longitude -24o). It describes growth rate according to light and temperature, associated nutrient constraints (light, nitrogen, and phosphorus), and acclimation consequences (glycogen and lipid productions per biomass) resumed by the glycogen storage index (see Material and Methods for details), with blue colors indicating consumption of potential glycogen stock and green colors showing increased storage. b |Distribution of lipid contribution to Prochlorococcus MED4 biomass over five distinct temperature ranges (see Materials and Methods for details). c |Distribution of glycogen production satisfying predicted Prochlorococcus MED4 growth rates under four categories of gradual light exposures. d |Distribution of glycogen storage indices computed with estimated Prochlorococcus MED4 growth rates under four similar categories of gradual light exposures. Indices between 0 and 0.46 index indicate a gradual decrease of glycogen stocks to support growth. Above 0.46, indices are associated with full phototrophic growth with increased glycogen storage. — biorxiv.org
https://www.biorxiv.org/content/10.1101/2023.11.23.568447v1
Towards modeling genome-scale knowledge in the global ocean (open access)
Astrobiology , Genomics,