Expanding The Natural Genetic Code – Implications For SynBio And Origin Of Life?
Editor’s note: The genetic code of all life (with a few minor exceptions) on our home world is based on a genomic code consisting of 4 standard nucleotides. It is possible that Earth life may have once used a different assortment of letters in its instructional alphabet but the evidence suggests that the current code has been in use for quite some time. But the genomics of life on other worlds might be different and use a different genetic alphabet. Also, as we tinker with earthly genomics for commercial and health reasons we rae discovering novel ways to tweak the standard genomic model to alter the outcome of a genetic sequence. This study using the Artificially Expanded Genetic Information Systems (AEGIS) shows that the pairing of non-standard nucleotides is at least possible. Whether the new sequences will work is another matter. But it does give us insight into how genetic sequences work and, by extension, how they might work elsewhere.
______________
Artificially Expanded Genetic Information Systems (AEGIS) add independently replicable unnatural nucleotide pairs to the natural G:C and A:T/U pairs found in native DNA, joining the unnatural pairs through alternative modes of hydrogen bonding. Whether and how AEGIS pairs are recognized and processed by multi-subunit cellular RNA polymerases (RNAPs) remains unknown.
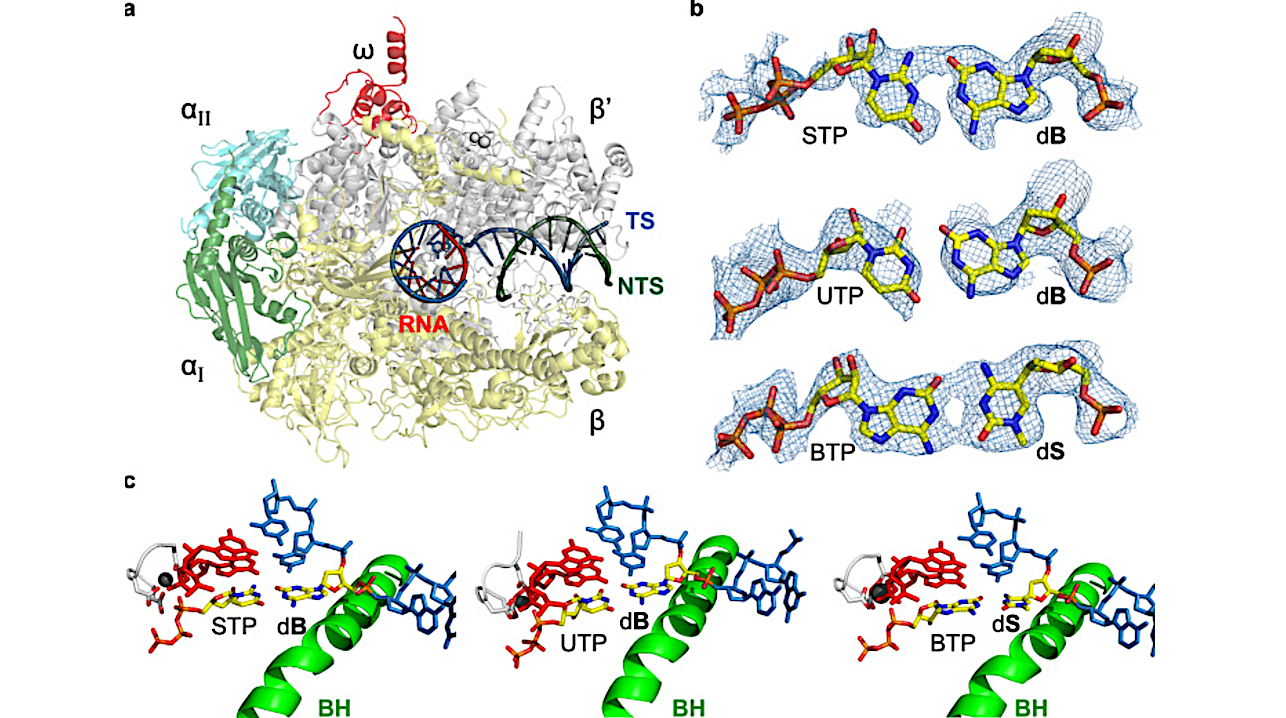
Here, we show that E. coli RNAP selectively recognizes unnatural nucleobases in a six-letter expanded genetic system. High-resolution cryo-EM structures of three RNAP elongation complexes containing template-substrate UBPs reveal the shared principles behind the recognition of AEGIS and natural base pairs.
In these structures, RNAPs are captured in an active state, poised to perform the chemistry step. At this point, the unnatural base pair adopts a Watson-Crick geometry, and the trigger loop is folded into an active conformation, indicating that the mechanistic principles underlying recognition and incorporation of natural base pairs also apply to AEGIS unnatural base pairs.
These data validate the design philosophy of AEGIS unnatural basepairs. Further, we provide structural evidence supporting a long-standing hypothesis that pair mismatch during transcription occurs via tautomerization. Together, our work highlights the importance of Watson-Crick complementarity underlying the design principles of AEGIS base pair recognition.
Juntaek Oh, Zelin Shan, Shuichi Hoshika, Jun Xu, Jenny Chong, Steven A. Benner, Dmitry Lyumkis & Dong Wang
A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase, Nature Communications (open access)
Astrobiology, Genomics, SynBio,