Bacteriophage-host Interactions In Microgravity Onboard The International Space Station
Bacteriophage-host interactions play a fundamental role in shaping microbial ecosystems. Although phage-host interactions have been extensively studied in terrestrial ecosystems, the impact of microgravity on phage-host interactions is yet to be investigated.
Here, we report the dynamics of interactions between T7 bacteriophage and E. coli in microgravity onboard the International Space Station (ISS). We found phage activity was delayed but ultimately successful in microgravity.
We identified several de novo mutations in phage and bacteria that improved fitness in microgravity. Deep mutational scanning of the receptor binding domain revealed substantial differences in the number, position, and mutational preferences between gravity and microgravity, reflecting underlying differences in the bacterial adaptations.
Combinatorial libraries informed by microgravity selections gave T7 mutants with 100-10,000-fold higher activity on uropathogenic E. coli under terrestrial conditions than wildtype T7. Our findings lay the foundation for future research on the impact of microgravity on phage-host interactions and microbial communities.
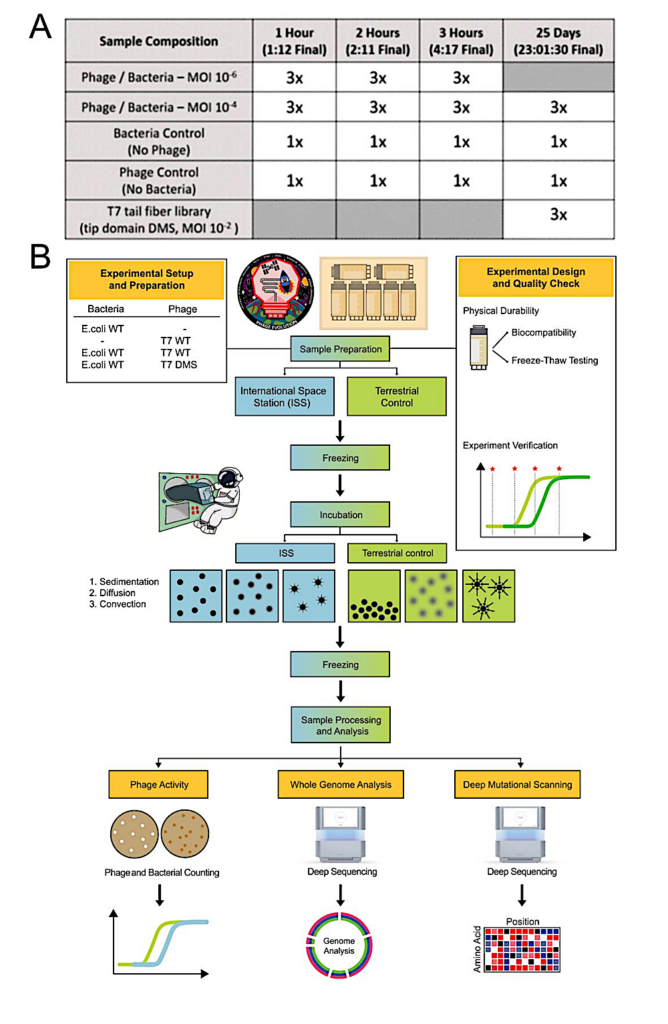
Experimental design to evaluate microgravity interactions on the ISS (A) Summary of samples and timepoints used in experiments. Final time in parenthesis indicates actual incubation time on the ISS, which was subsequently used for terrestrial controls. (B) Illustration of experimental process to prepare and test samples under terrestrial conditions and in microgravity onboard the International Space Station (ISS). Samples were prepared on Earth, and quality checked to ensure samples cannot leak and cryovial integrity is unaffected by the freeze-thaw process. Two sets of samples were tested, one in microgravity in the ISS (left) and one on Earth (right). Samples were frozen, then thawed and incubated in either condition for specified intervals. Samples were then re-frozen and evaluated on Earth for phage and bacterial counts, for whole genome sequencing, and for deep mutational scanning of the T7 receptor binding protein tip domain. Image courtesy of NASA/Rhodium Scientific. — biorxiv

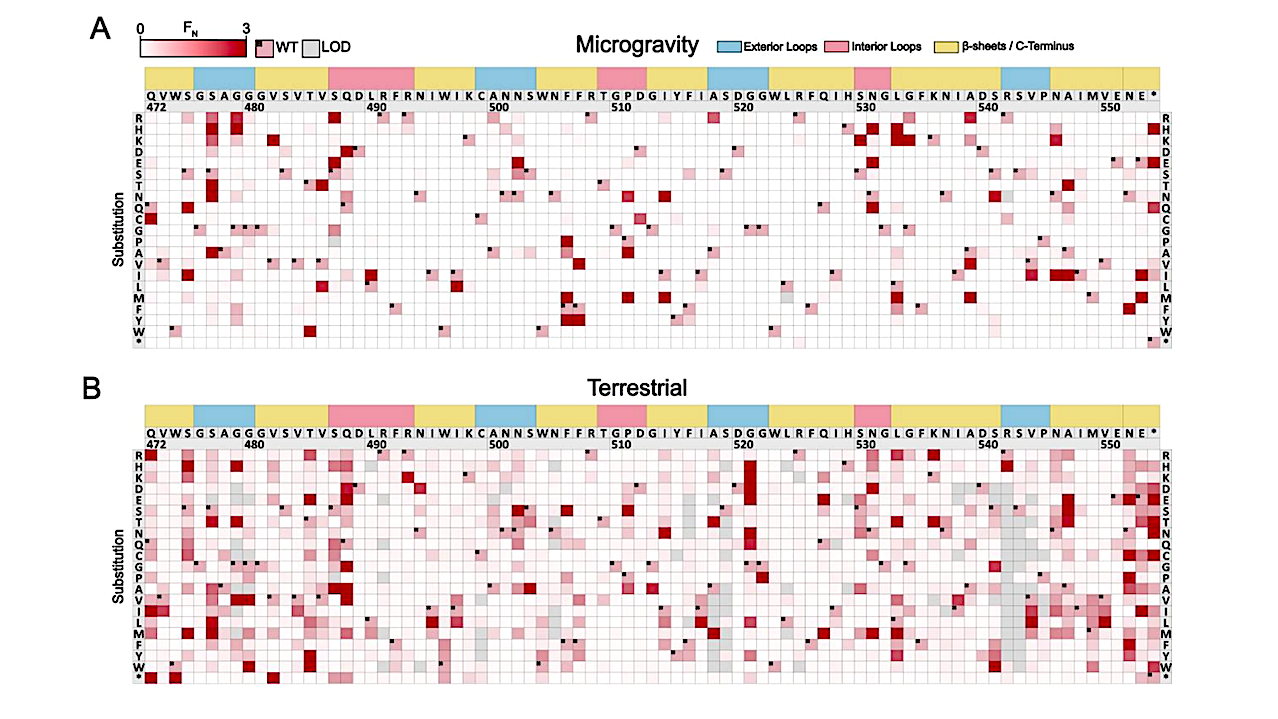
Deep Mutational Scanning profiles beneficial substitutions in the RBP tip domain (A-B) Heatmaps showing normalized functional scores (FN) of all substitutions (red gradient) and wildtype amino acids (FN=1 and black dot upper left) at every position after incubation with E. coli BL21 in (A) microgravity and (B) after terrestrial incubation. Residue numbering (based on PDB 4A0T), wildtype amino acid and secondary structure topology (exterior loops in blue, interior loops in red, beta sheets and C-terminus in yellow) are shown above left to right, substitutions listed top to bottom. Substitutions below limit of detection (LOD) where a score cannot be called are shown in yellow. (C) Correlation plot of log2 transformed FN after microgravity (purple) and terrestrial (green) incubation with example divergent substitutions colored accordingly. (D) Tip domain crystal structure (PDB: 4A0T) and secondary structure topology shown color coded with noted substitutions enriched in microgravity (purple) or terrestrial (green) incubation conditions. (E) Proportion of enriched substitutions based on total FN by topological location throughout the tip domain in microgravity (purple) or terrestrial (green) incubation conditions. — biorxiv
Bacteriophage-host interactions in microgravity onboard the International Space Station
https://www.biorxiv.org/content/10.1101/2023.10.10.561409v2
Astrobiology