Artificial Intelligence Tools Shed Light On Millions Of Proteins

Editor’s note: life on Earth uses a plethora of proteins. But there are many more proteins – millions – that can exist – many, many more than are used by terrestrial biology. What proteins will be used in the metabolism of life forms on other worlds? Will they be very similar to us, slightly similar, or totally ‘alien’?
______________
A research team at the University of Basel and the SIB Swiss Institute of Bioinformatics uncovered a treasure trove of uncharacterised proteins. Embracing the recent deep learning revolution, they discovered hundreds of new protein families and even a novel predicted protein fold. The study has now been published in Nature.
In the past years, AlphaFold has revolutionised protein science. This Artificial Intelligence (AI) tool was trained on protein data collected by life scientists for over 50 years, and is able to predict the 3D shape of proteins with high accuracy. Its success prompted the modelling of an astounding 215 million proteins last year, providing insights into the shapes of almost any protein. This is particularly interesting for proteins that have not been studied experimentally, a complex and time-consuming process.
“There are now many sources of protein information, enclosing valuable insights into how proteins evolve and work” says Joana Pereira, the leader of the study. Nevertheless, research has long been faced with a data jungle. The research team led by Professor Torsten Schwede, group leader at the Biozentrum, University of Basel, and the Swiss Institute of Bioinformatics (SIB), has now succeeded in decrypting some of the concealed information.
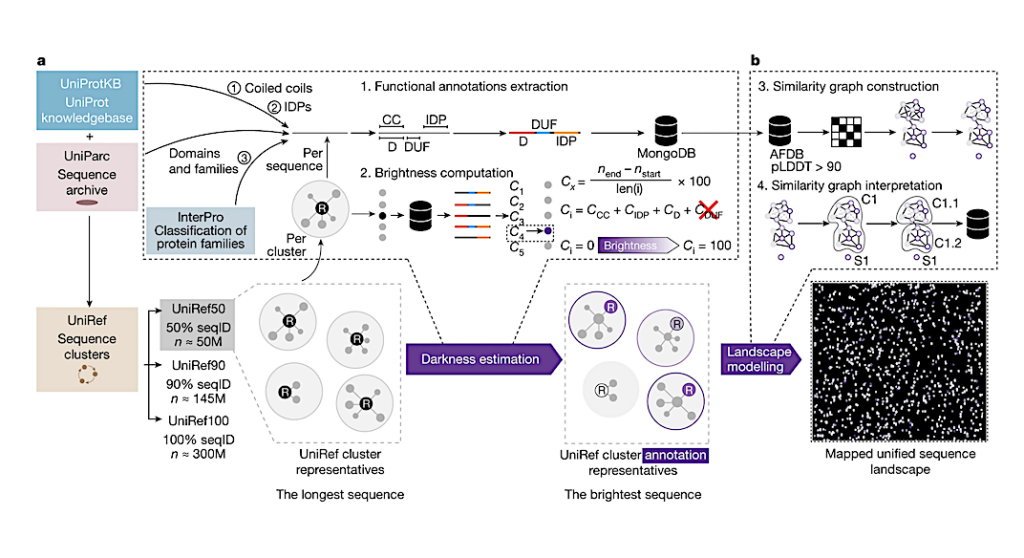
a, Starting from the clusters in UniRef50, we collected all the functional annotations for all included UniProtKB and UniParc entries, including domain (D), coiled-coil (CC) and intrinsically disordered (IDPs) predictions and excluding all of those with putative, hypothetical, uncharacterized and DUF in their names. Cx corresponds to the coverage of an annotation, Ci corresponds to the functional brightness across the entire sequence. We selected the protein with the highest full-length annotation coverage (that is, brightness, Ci) as the functional representative of each cluster. b, From the collected UniRef50 clusters, we selected those with a structural representative with pLDDT greater than 90 in the AFDB v.4, and constructed a large-scale sequence similarity network by all-against-all MMseqs2 searches, representing the sequence landscape of more than 6 million UniRef50 clusters. — Nature
A bird’s eye view reveals new protein families and folds
The researchers constructed an interactive network of 53 million proteins with high quality AlphaFold structures. “This network serves as a valuable source for theoretically predicting unknown protein families and their functions on a large scale,” underlines Dr. Janani Durairaj, the first author. The team was able to identify 290 new protein families and one new protein fold that resembles the shape of a flower.
Building on the expertise of the Schwede group in developing and maintaining the leading software SWISS-MODEL, they made the network available as an interactive web resource, termed the “Protein Universe Atlas”.
AI as a valuable tool in research
The team has employed Deep Learning-based tools for finding novelties in this network, paving the way to innovations in life sciences, from basic to applied research. “Understanding the structure and function of proteins is typically one of the first steps to develop a new drug, or modify their functions by protein engineering, for example”, says Pereira. The work was supported by a ‘kickstarter’ grant from SIB to encourage the adoption of AI in life science resources. It underscores the transformative potential of Deep Learning and intelligent algorithms in research.
With the Protein Universe Atlas, scientists can now learn more about proteins relevant to their research. “We hope this resource will help not only researchers and biocurators but also students and teachers by providing a new platform for learning about protein diversity, from structure, to function, to evolution”, says Janani Durairaj.
Protein Universe Atlas: https://uniprot3d.org/atlas/AFDB90v4
Uncovering new families and folds in the natural protein universe, Nature (open access)
Astrobiology