Tricorder Tech: Translation As A Biosignature
Life on Earth relies on mechanisms to store heritable information and translate this information into cellular machinery required for biological activity. In all known life, storage, regulation, and translation are provided by DNA, RNA, and ribosomes.
Life beyond Earth, even if ancestrally or chemically distinct from life as we know it may utilize similar structures: it has been proposed that charged linear polymers analogous to nucleic acids may be responsible for storage and regulation of genetic information in non-terran biochemical systems. We further propose that a ribosome-like structure may also exist in such a system, due to the evolutionary advantages of separating heritability from cellular machinery.
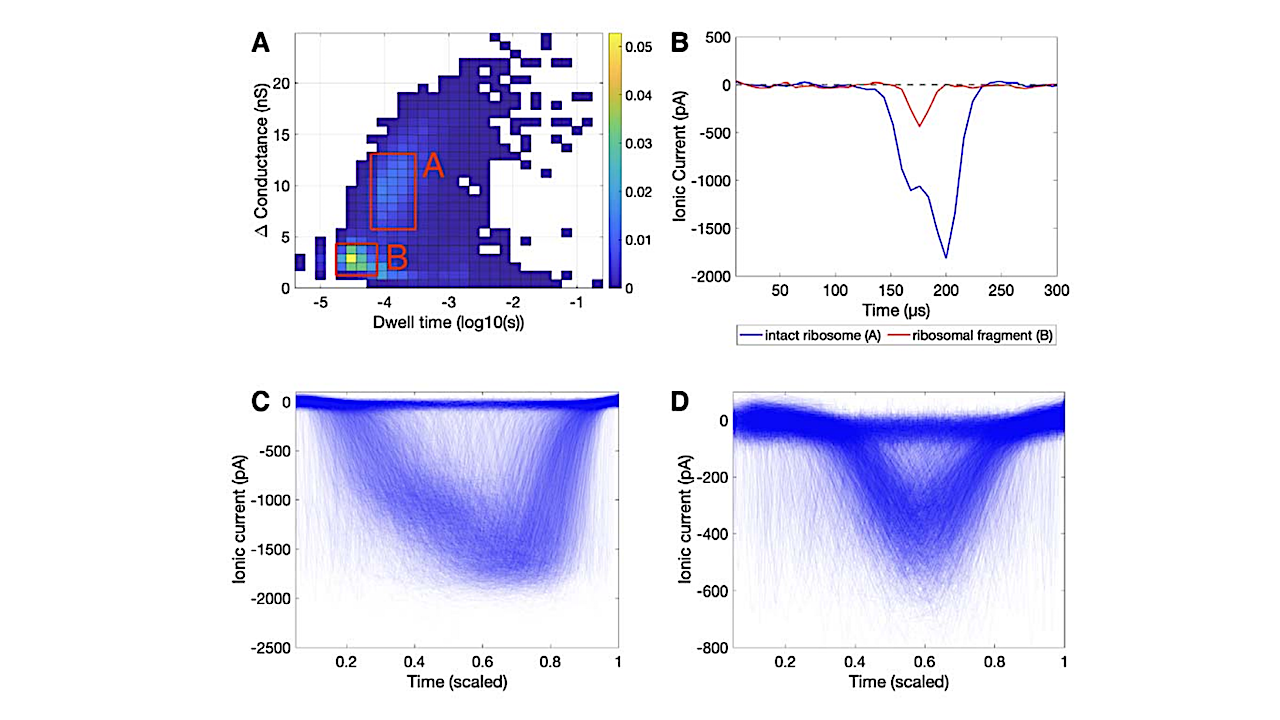
Here, we use a solid-state nanopore to detect DNA, RNA, and ribosomes, and demonstrate that machine learning can distinguish between biomolecule samples and accurately classify new data. This work is intended to serve as a proof of principal that such biosignatures (i.e., informational polymers or translation apparatuses) could be detected, for example, as part of future missions targeting extant life on Ocean Worlds. A negative detection does not imply the absence of life; however, detection of ribosome-like structures could provide a robust and sensitive method to seek extant life in combination with other methods.
doi: https://doi.org/10.1101/2023.08.10.552839
https://www.biorxiv.org/content/10.1101/2023.08.10.552839v2, biorxiv.org
Astrobiology