Earth’s Soil Microbiome Is Under Threat From Climate Change
Using a novel method to detect microbial activity in biological soil crusts, or biocrusts, after they are wetted, a Penn State-led research team in a new study uncovered clues that will lead to a better understanding of the role microbes play in forming a living skin over many semi-arid ecosystems around the world. The tiny organisms — and the microbiomes they create — are threatened by climate change.
The researchers published their findings in Frontiers of Microbiology.
“Biocrusts currently cover approximately 12% of Earth’s terrestrial surface, and we expect them to decrease by about 25% to 40% within 65 years due to climate change and land-use intensification,” said team leader Estelle Couradeau, Penn State assistant professor of soils and environmental microbiology. “We hope this work can pave the way to understanding the microbial functions supporting biocrust resilience to the rapidly changing climate patterns and more frequent droughts.”
Biological soil crusts are assemblages of organisms that form a perennial, well-organized surface layer in soils. They are widespread, occurring on all of the continents wherever a shortage of water limits the growth of common plants, allowing light to reach bare soil. But there is still sufficient water to support the growth of microorganisms that perform valuable ecosystem services such as taking carbon and nitrogen from the air and fixing them in the soil, recycling nutrients and holding soil particles together, which helps prevent dust.
That soil-stabilizing function — which reduces erosion by providing the means for soil to clump and not break down into dust — is extremely important, according to Couradeau. Her research group, now in Penn State’s College of Agricultural Sciences, has been intensively studying biocrusts for a decade.
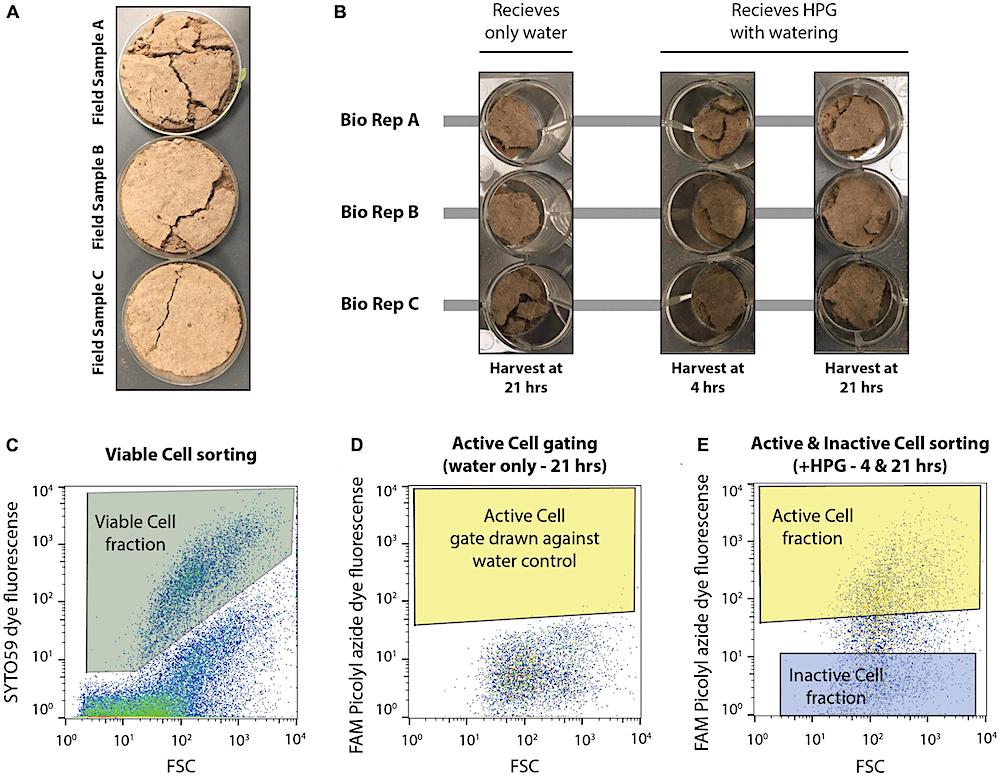
Experimental set up of microcosms and design of flow cytometry-based cell sorting. (A) Three biological replicate field samples of biocrusts were collected from Moab, UT in petri dishes. (B) Field samples were distributed to a total of nine wells of a 12 well plate. Each biological replicate (bio rep) was distributed to three wells each. This first set of biocrusts received only water during the simulated rain event and was harvested 21 hrs later. The remaining six wells received HPG during soil wetting and were harvested at either 4 or 21 hrs after incubation. (C) SYTO59 fluorescence was used to collect intact, viable cells (Viable Cell fraction), discriminating these particles from non-stained particles. (D) Microcosms receiving only water were used to identify the background FAM Picolyl azide dye fluorescence. Fluorescence above this background (giving a false positive rate of >0.05%) represented active cells (Active Cell gate) and any particles falling within both the Viable Cell gate and the Active Cell gate were assigned to the Active Cell fraction. (E) The Inactive Cell gate was drawn at least a half an order of magnitude lower in FAM Picolyl azide dye fluorescence, and cells within the Viable Cell gate and Inactive Cell gate were assigned to the Inactive Cell fraction when sorting, while cells falling within the Viable Cell and Active Cell gate were assigned to the Active Cell fraction. Cell sorting was performed on microcosms receiving HPG and harvested at either 4 or 21 hrs following the wetting event.
“Most dust is generated in drylands, and studies suggest that the presence of biocrusts in drylands greatly reduce the amount of dust that would otherwise make its way into the atmosphere,” she said. “We think losing biocrusts would cause a 5% to 15% increase in global dust emission and deposition — which would affect the climate, environment and human health.”
In the semi-arid regions where biocrusts exist, the organisms — tiny mosses, lichens, green algae, cyanobacteria, other bacteria and fungi — may experience just a few rain or snow events a year, explained Ryan Trexler, a doctoral degree candidate in the Intercollege Graduate Degree Program in ecology and in biogeochemistry, who spearheaded the research.
“When the soil is dry, for the most part, the microbes in the soil are dormant, not doing much,” he said. “But as soon as they sense water, they’re resuscitated very quickly, within seconds to minutes. And they are actively making chlorophyll and fixing carbon and nitrogen until the soil is dry again — and then the microbes go dormant again. They go through cycles of activity every time it rains.”
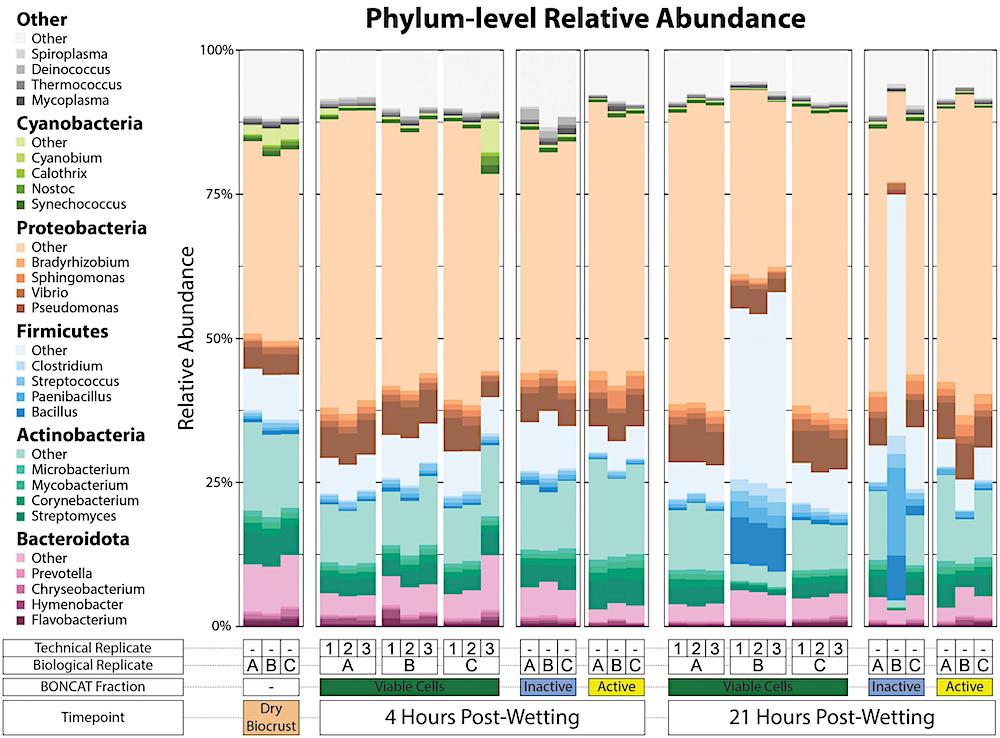
Phylum-level relative abundance of kraken2/bracken taxonomy output. The top five most abundant phyla across the dataset are plotted as five different color pallets and the top four genera of each of these phyla are filled with various shades of the phylum color pallet. Other top genera are filled with shades of gray. A key for the metagenome sample type is included below the x-axis.
To study biocrusts, the researchers took samples from three plots of undisturbed, cyanobacteria-dominated biocrusts located on the Colorado Plateau near Moab, Utah. Biocrust samples were taken in fall following rain that wetted the soil sufficiently to activate the microbes. The samples were subsequently dried and stored in the dark and then rewetted much later in the research.
“We sampled what we call ‘a cold desert,’ because it’s very arid, but in the winter, it sometimes snows,” Trexler said. “So, it’s not as hot as many other arid places, but still plants cannot thrive there because there’s not enough water. And so, the only community that we find in soils at the site are microbial.”
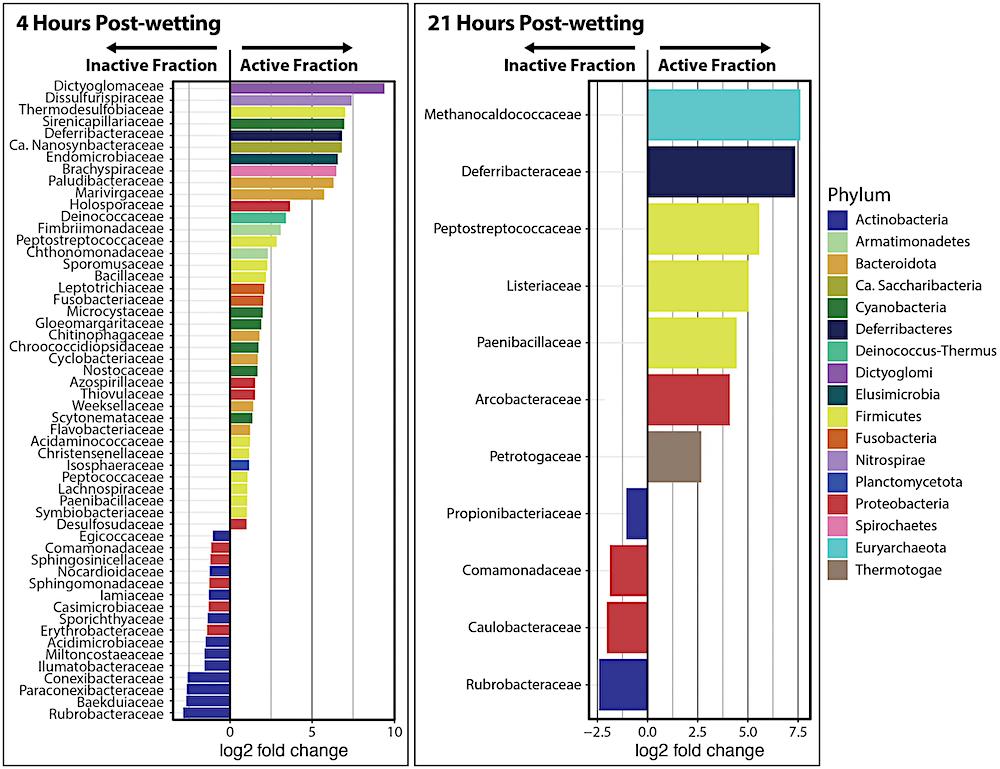
Differential abundance analysis of kraken2/bracken taxa at the family level at 4 and 21 hrs after biocrust wetting. Only significantly different families between the Inactive Cell and Active Cell fractions are plotted (α = 0.05). A positive log2 fold change value indicates higher abundance in the Active Cell fraction while a negative log2 fold change value represents a higher abundance in the Inactive Cell fraction. Barplot colors represent the phylum in which each family belongs.
To determine which microorganisms are active within soil communities, the researchers coupled bioorthogonal non-canonical amino acid tagging — known as BONCAT — with fluorescence-activated cell sorting. BONCAT is a powerful tool for tracking protein synthesis on the level of single cells within communities and whole organisms, while fluorescence-activated cell sorting sorts cells based on whether they are producing new proteins.
The researchers combined these processes with shotgun metagenomic sequencing, which allowed them to comprehensively sample all genes in all organisms present in biocrust samples. They applied this method to profile the diversity and potential functional capabilities of both active and inactive microorganisms in a biocrust community after being resuscitated by a simulated rain event. The researchers found that their novel approach can discern active and inactive microorganisms in wetted biocrusts.
The active and inactive components of the biocrust community differed in species richness and composition at both four hours and 21 hours after the wetting event, the researchers reported.
Contributing to the research were Marc Van Goethem, Lawrence Berkeley National Laboratory, and King Abdullah University of Science and Technology, Jeddah, Saudi Arabia; Danielle Goudeau, Nandita Nath, Trent Northen and Rex Malmstrom, Lawrence Berkeley National Laboratory, U.S. Department of Energy Joint Genome Institute.
The U.S. Department of Energy supported this research.
BONCAT-FACS-Seq reveals the active fraction of a biocrust community undergoing a wet-up event
Astrobiology