Artificial Cells Demonstrate That “Life Finds A Way”
“Listen, if there’s one thing the history of evolution has taught us is that life will not be contained. Life breaks free. It expands to new territories, and it crashes through barriers painfully, maybe even dangerously, but . . . life finds a way,” said Ian Malcolm, Jeff Goldblum’s character in Jurassic Park, the 1993 science fiction film about a park with living dinosaurs.
“Listen, if there’s one thing the history of evolution has taught us is that life will not be contained. Life breaks free. It expands to new territories, and it crashes through barriers painfully, maybe even dangerously, but . . . life finds a way,” said Ian Malcolm, Jeff Goldblum’s character in Jurassic Park, the 1993 science fiction film about a park with living dinosaurs.
You won’t find any Velociraptors lurking around evolutionary biologist Jay T. Lennon’s lab; however, Lennon, a professor in the College of Arts and Sciences Department of Biology at Indiana University Bloomington, and his colleagues have found that life does indeed find a way. Lennon’s research team has been studying a synthetically constructed minimal cell that has been stripped of all but its essential genes. The team found that the streamlined cell can evolve just as fast as a normal cell—demonstrating the capacity for organisms to adapt, even with an unnatural genome that would seemingly provide little flexibility.
“It appears there’s something about life that’s really robust,” says Lennon. “We can simplify it down to just the bare essentials, but that doesn’t stop evolution from going to work.”
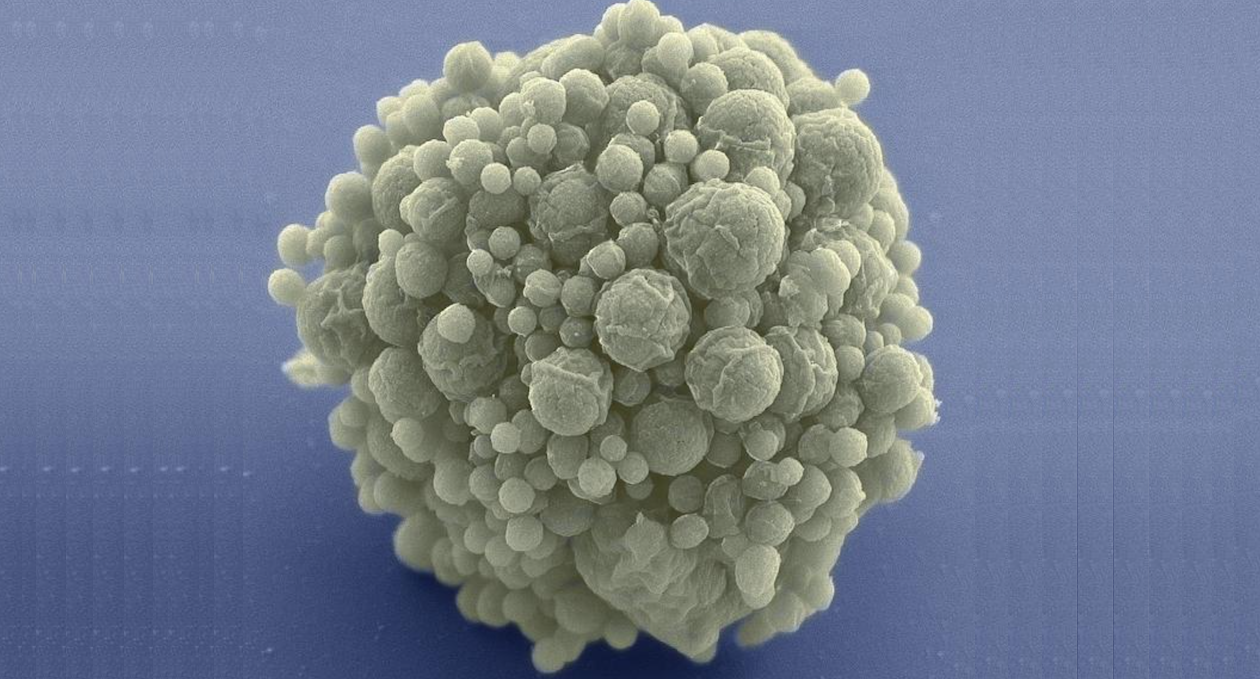
For their study, Lennon’s team used the synthetic organism, Mycoplasma mycoides JCVI-syn3B—a minimized version of the bacterium M. mycoides commonly found in the guts of goats and similar animals. Over millennia, the parasitic bacterium has naturally lost many of its genes as it evolved to depend on its host for nutrition. Researchers at the J. Craig Venter Institute in California took this one step further. In 2016, they eliminated 45 percent of the 901 genes from the natural M. mycoides genome—reducing it to the smallest set of genes required for autonomous cellular life. At 493 genes, the minimal genome of M. mycoides JCVI-syn3B is the smallest of any known free-living organism. In comparison, many animal and plant genomes contain more than 20,000 genes.
In principle, the simplest organism would have no functional redundancies and possess only the minimum number of genes essential for life. Any mutation in such an organism could lethally disrupt one or more cellular functions, placing constraints on evolution. Organisms with streamlined genomes have fewer targets upon which positive selection can act, thus limiting opportunities for adaptation.
Although M. mycoides JCVI-syn3B could grow and divide in laboratory conditions, Lennon and colleagues wanted to know how a minimal cell would respond to the forces of evolution over time, particularly given the limited raw materials upon which natural selection could operate as well as the uncharacterized input of new mutations.
“Every single gene in its genome is essential,” says Lennon in reference to M. mycoides JCVI-syn3B. “One could hypothesize that there is no wiggle room for mutations, which could constrain its potential to evolve.”
The researchers established that M. mycoides JCVI-syn3B, in fact, has an exceptionally high mutation rate. They then grew it in the lab where it was allowed to evolve freely for 300 days, equivalent to 2000 bacterial generations or about 40,000 years of human evolution.
The next step was to set up experiments to determine how the minimal cells that had evolved for 300 days performed in comparison to the original, non-minimal M. mycoides as well as to a strain of minimal cells that hadn’t evolved for 300 days. In the comparison tests, the researchers put equal amounts of the strains being assessed together in a test tube. The strain better suited to its environment became the more common strain.
They found that the non-minimal version of the bacterium easily outcompeted the unevolved minimal version. The minimal bacterium that had evolved for 300 days, however, did much better, effectively recovering all of the fitness that it had lost due to genome streamlining. The researchers identified the genes that changed the most during evolution. Some of these genes were involved in constructing the surface of the cell, while the functions of several others remain unknown.
Details about the study can be found in a paper recently featured in Nature. Roy Z. Moger-Reischer, a Ph.D. student in the Lennon lab at the time of the study, is first author on the paper.
Understanding how organisms with simplified genomes overcome evolutionary challenges has important implications for long-standing problems in biology—including the treatment of clinical pathogens, the persistence of host-associated endosymbionts, the refinement of engineered microorganisms, and the origin of life itself. The research done by Lennon and his team demonstrates the power of natural selection to rapidly optimize fitness in the simplest autonomous organism, with implications for the evolution of cellular complexity. In other words, it shows that life finds a way.
Evolution of a minimal cell, Nature (open access)
Astrobiology