Tricorder Tech: Developing An Astrobiological Nomenclature Code For Offworld Use
Editor’s note: The article “SeqCode: a nomenclatural code for prokaryotes described from sequence data” recently appeared in the journal Nature Microbiology. The article deals with a process (SeqCode) whereby genomic sequence data is used in the preparation of nomenclature used to name and define an organism – in this case prokaryotes. With regard to Astrobiology, how will we map out the genomic relationships of extraterrestrial life – and name these life forms in an orderly, logical fashion? Perhaps SeqCode can be used as a model for such a process.
Abstract source: PubMed citation.
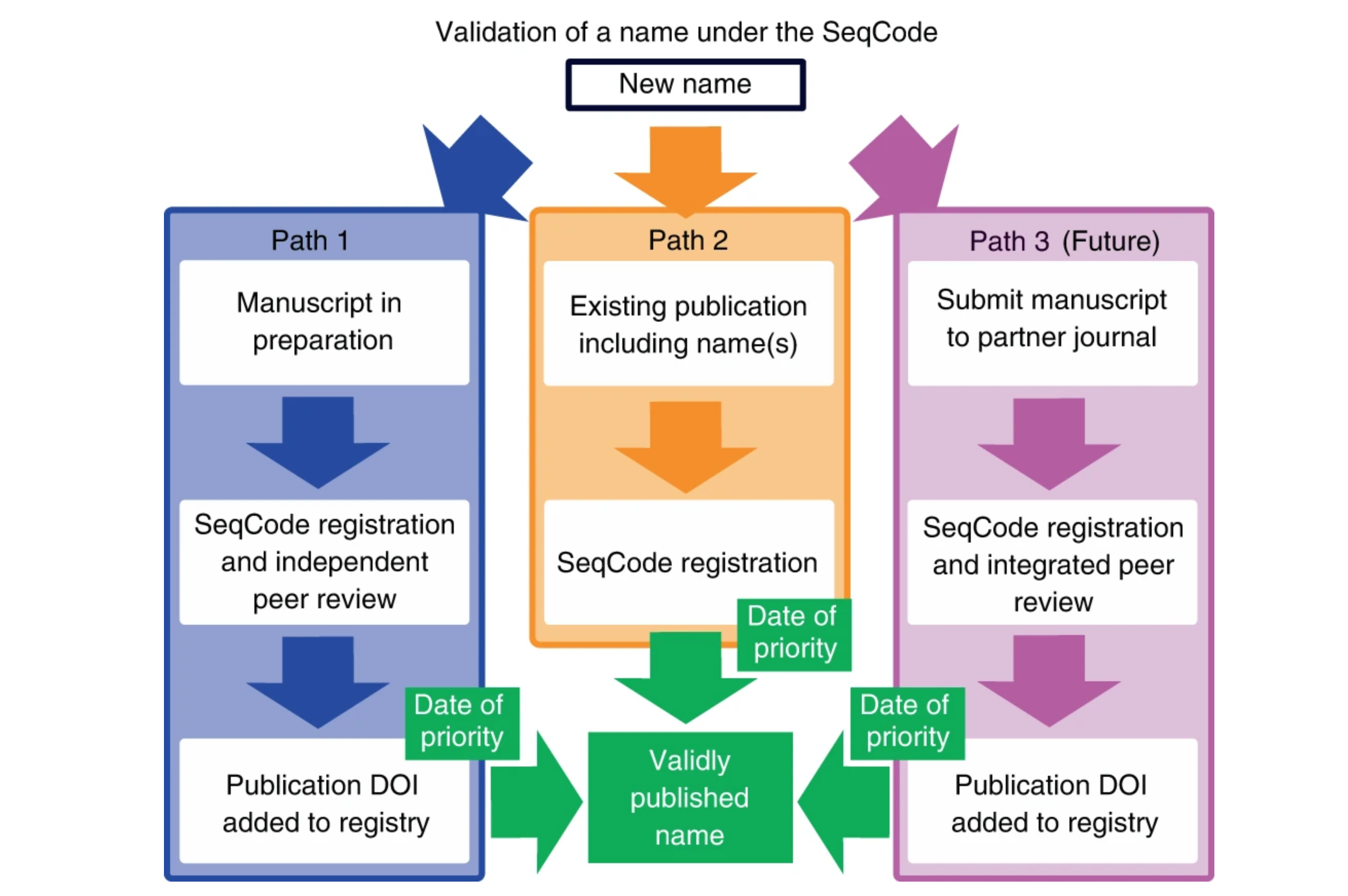
“Validation process of a name under the SeqCode. Currently, two mechanisms exist, with a third possible in the future. Hedlund et al, Nature Microbiology: Most prokaryotes are not available as pure cultures and therefore ineligible for naming under the rules and recommendations of the International Code of Nomenclature of Prokaryotes (ICNP).
Here we summarize the development of the SeqCode, a code of nomenclature under which genome sequences serve as nomenclatural types. This code enables valid publication of names of prokaryotes based upon isolate genome, metagenome-assembled genome or single-amplified genome sequences.
Otherwise, it is similar to the ICNP with regard to the formation of names and rules of priority. It operates through the SeqCode Registry (https://seqco.de/), a registration portal through which names and nomenclatural types are registered, validated and linked to metadata.
We describe the two paths currently available within SeqCode to register and validate names, including Candidatus names, and provide examples for both. Recommendations on minimal standards for DNA sequences are provided. Thus, the SeqCode provides a reproducible and objective framework for the nomenclature of all prokaryotes regardless of cultivability and facilitates communication across microbiological disciplines.”
Full article: SeqCode: a nomenclatural code for prokaryotes described from sequence datam Nature Microbiology
Full caption: “Validation process of a name under the SeqCode. Currently, two mechanisms exist, with a third possible in the future. In the recommended mechanism (blue arrows, path 1), draft registration of the name and entry of metadata into the SeqCode Registry occurs concurrently with preparation of the effective publication. Within the Registry, data quality and name synonymy checks in conjunction with curator review take place as described in Tables 2 and 3, leading to provisional acceptance of proposals that comply with SeqCode rules. This procedure ensures data quality and avoids requiring errata after publication for corrections. Entry of the DOI of the publication into the Registry marks the time and date of priority. Because the SeqCode requires that the earliest name of a taxon be used, the date of priority establishes the precedence of this name as the only valid name for the taxon. The second (orange arrows, path 2) is for names that are already published, such as Candidatus names. The name and metadata are entered into the Registry. Automated checks and SeqCode curators review compliance and acceptance of the proposal completes registration and marks the time and date of priority. At that point, the Candidatus designation can be removed. The third mechanism could be developed in partnership with one or more journals in the future (pink arrows, path 3). It would involve simultaneous peer review and Registry curator review as an integrated path to the validation of proposed names. Issuance of the DOI of the accepted paper marks the time and date of priority. Please see the text and Supplementary Information for concrete examples of registration through paths 1 and 2.”
Astrobiology, Genomics,