Tricorder Tech: A Universal Sequencing Read Interpreter – INTERSTELLAR (interpretation, scalable transformation, and emulation of large-scale sequencing reads)
Massively parallel DNA sequencing has led to the rapid growth of highly multiplexed experiments in biology. Such experiments produce unique sequencing results that require specific analysis pipelines to decode highly structured reads.
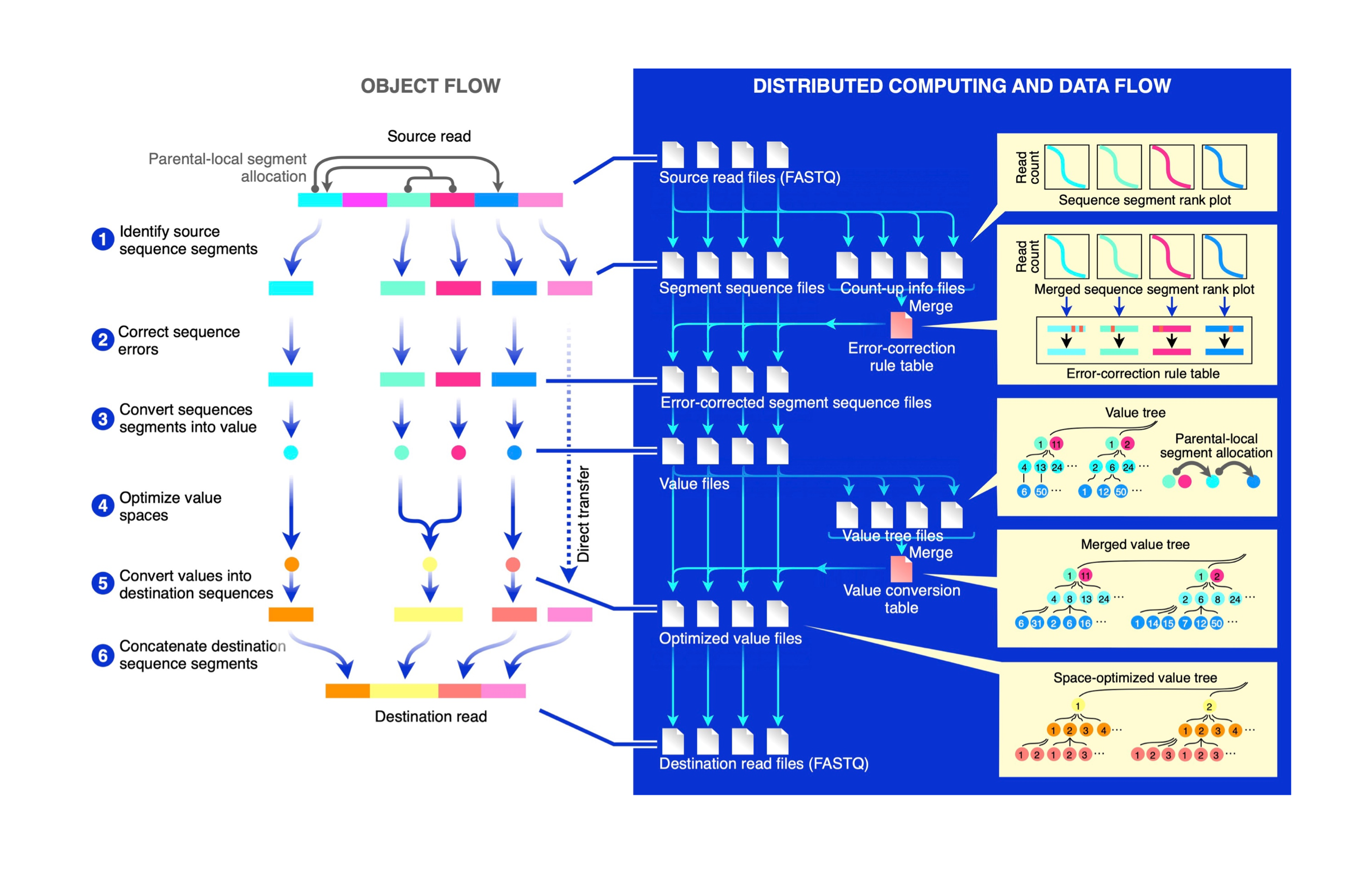
However, no versatile framework that interprets sequencing reads for downstream biological analysis has been developed. Here we report INTERSTELLAR (interpretation, scalable transformation, and emulation of large-scale sequencing reads) that extracts data values encoded in theoretically any type of sequencing read and translates them into sequencing reads of any structure of choice.
INTERSTELLAR first identifies sequence segments encoded in reads according to the user’s definition. Followed by error correction, values are extracted and compressed into an optimal space, which can be efficiently translated into sequencing reads of another structure.
We demonstrated that INTERSTELLAR successfully extracted information from a range of sequencing reads and translated those of single-cell (sc)RNA-seq, scATAC-seq, and spatial transcriptomics to be analyzed by different software tools that have been developed for conceptually the same types of experiments.
INTERSTELLAR will greatly facilitate the development of new sequencing-based experiments and sharing of data analysis pipelines.
PDF of full paper
Yusuke Kijima, Daniel Evans-Yamamoto, Hiromi Toyoshima, Nozomu Yachie
doi: https://doi.org/10.1101/2022.04.16.488535
Astrobiology, Genomics, Tricorder,