Extremophile Dye Labeling And Searching For Life On Mars

Researchers from MIPT and their colleagues from Research Center Juelich (Germany) and Dmitry Mendeleev University of Chemical Technology of Russia have described a new method for studying microorganisms that can survive in extreme conditions.
The scientists identified a fluorescent dye that enabled them to observe the life cycle of bacteria in real time.
Halophiles, which is the ancient Greek for “salt-loving,” are microorganisms that thrive in high salt concentrations. Their ability to survive in hostile environments makes halophiles important scientific objects, for both theoretical and applied studies. This line of enquiry may eventually facilitate the search for extraterrestrial life, shed light on the history of the Earth, and provide data sought by biotech specialists. The authors of the paper, who work at MIPT’s Laboratory for Advanced Studies of Membrane Proteins, point out that these organisms can be used for many purposes, including cleaning up oil spills. However, this research is facing a number of obstacles, not least of them being that microbiological experiments are technically quite challenging.
To study microorganisms in their natural environment, dyes are required — ideally, selective ones. With their help, much more data can be obtained, compared to when an unstained medium is examined. However, well-established fluorescent labels and antibodies that dyes use to bind with a given substance often fail to work in salty environments. Additional difficulties are posed by the halophiles’ thick membrane.
“Despite all the hard work, scientists have so far been unable to find a substance that would enable them to observe these organisms ‘live,’ the way they really are. Instead, bacteria had to undergo harmful preparation,” says Ivan Maslov, a fifth-year MIPT student and co-author of the study.
In the new paper published in Scientific Reports, the international research team described a solution to this issue. Their experiments showed that there is no need to synthesize new types of dyes: Previously created substances for labeling mitochondria in eukaryotic cells demonstrated positive results in halophiles as well.
Note: There are two major types of cells: prokaryotes and eukaryotes. Prokaryotes, represented by bacteria, lack nuclei, and other membrane-bound structures. Eukaryotes — animal, plant, and fungal cells — have nuclei and various organelles. Among them are mitochondria, which are used to generate adenosine triphosphate molecules — a universal energy source consumed in various cellular processes. Interestingly, the modern view on the subject suggests that mitochondria were originally free-living bacteria and only later became symbionts of eukaryotic cells. Even now, they still have their own DNA.
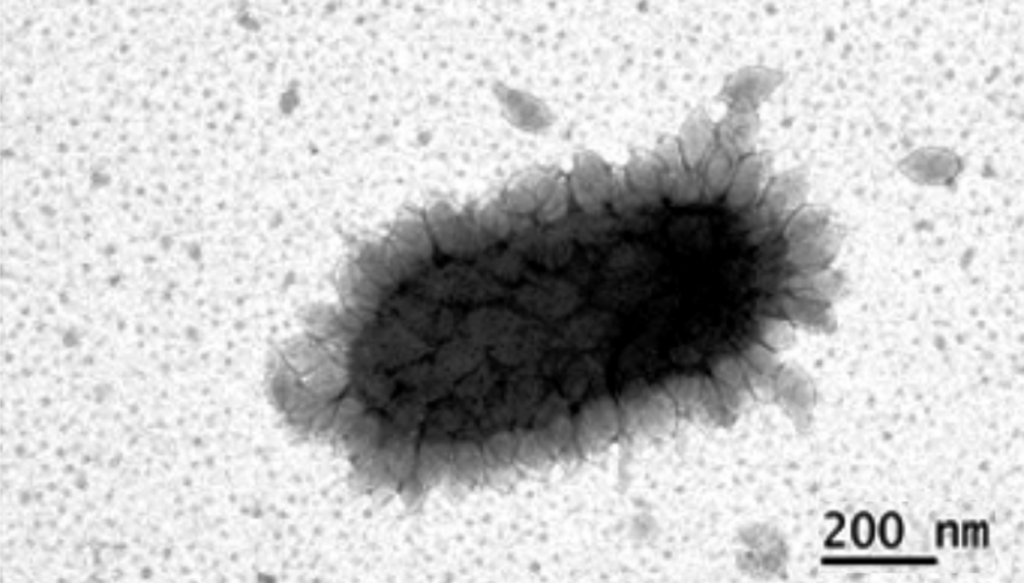
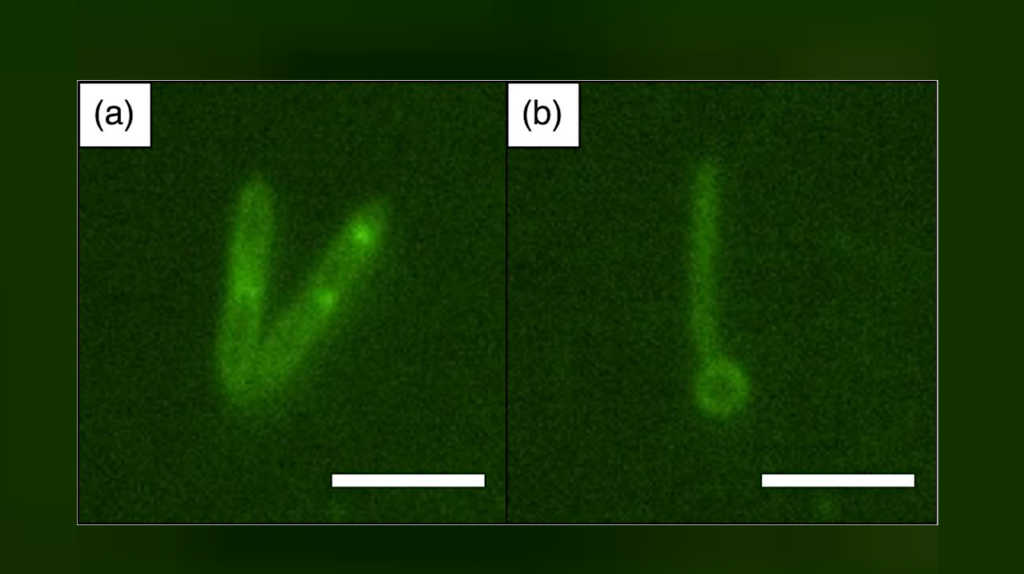
MitoTracker dyes proved to be successful in staining a wide range of microorganisms: Halobacterium salinarium, Haloferax sp., Halorubrum sp., Salicola sp., and Halomonas sp. (the letters “sp” mean “one of the species”). The experiments conducted by the researchers demonstrated that it is possible not only to obtain clear photos and keep count of the cells, but also to observe the transformation of Halobacterium salinarium. When exposed to hostile chemical treatment, the cells changed their shape: From rod-shaped, they turned into spheres. The team even made a video recording of that process.
The new method will be effective in labeling microorganisms in their natural environment, be it in a saline deposit on Earth or in a Martian soil sample retrieved by a rover. It will also help study the behavior of these bacteria with minimum distortions of the results.
“Halophiles are often found in ancient saline deposits that have been building up for millions of years. Our method helps locate these organisms in mineral formations and study them. This can shed light on the origin of life on Earth. According to one theory, life was brought to our planet from elsewhere in the form of bacteria,” comments Valentin Borshchevskiy, lead author of the study and deputy head of the Laboratory for Advanced Studies of Membrane Proteins, MIPT.
This research was supported by the Russian Science Foundation (16-15-00242) and the Ministry of Education and Science of the Russian Federation (personal support under project No. 6.9909.2017/??).