Tricorder Tech: Development Of Solid-State Nanopore Technology For Life Detection
Introduction: Biomarkers for life on Earth are an important starting point to guide the search for life elsewhere. However, the search for life beyond Earth should incorporate technologies capable of recognizing an array of potential biomarkers beyond what we see on Earth, in order to minimize the risk of false negatives from life detection missions.
With this in mind, charged linear polymers may be a universal signature for life, due to their ability to store information while also inherently reducing the tendency of complex tertiary structure formation that significantly inhibit replication. Thus, these molecules are attractive targets for biosignature detection as potential “self-sustaining chemical signatures” [1]. Examples of charged linear polymers, or polyelectrolytes, include deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) as well as synthetic polyelectrolytes that could potentially support life, including threose nucleic acid (TNA) and other xenonucleic acids (XNAs).
Nanopore analysis is a novel technology that has been developed for singlemolecule sequencing with exquisite single nucleotide resolution which is also well-suited for analysis of polyelectrolyte molecules. Nanopore analysis has the ability to detect repeating sequences of electrical charges in organic linear polymers, and it is not molecule-specific (i.e. it is not restricted to only DNA or RNA). In this sense, it is a better life detection technique than approaches that are based on specific molecules, such as the polymerase chain reaction (PCR), which requires that the molecule being detected be composed of DNA.
Rationale: Nanopore technology has been a revolutionary technology for nucleic acid sequencing [2]. The MinION, by Oxford Nanopore, has been used to sequence DNA in microgravity conditions for the first time ever, on board the International Space Station [3]. However, expecting current nanopore platforms to retain their functionality for long duration space missions is unrealistic. The robustness of the nanopores currently available commercially is limited by the stability of the membranes and modified nanopore proteins nanopores. Long duration space mission, involving life/biosignature detection, will need hardier technology to be developed; solid-state nanopores are an attractive alternative.
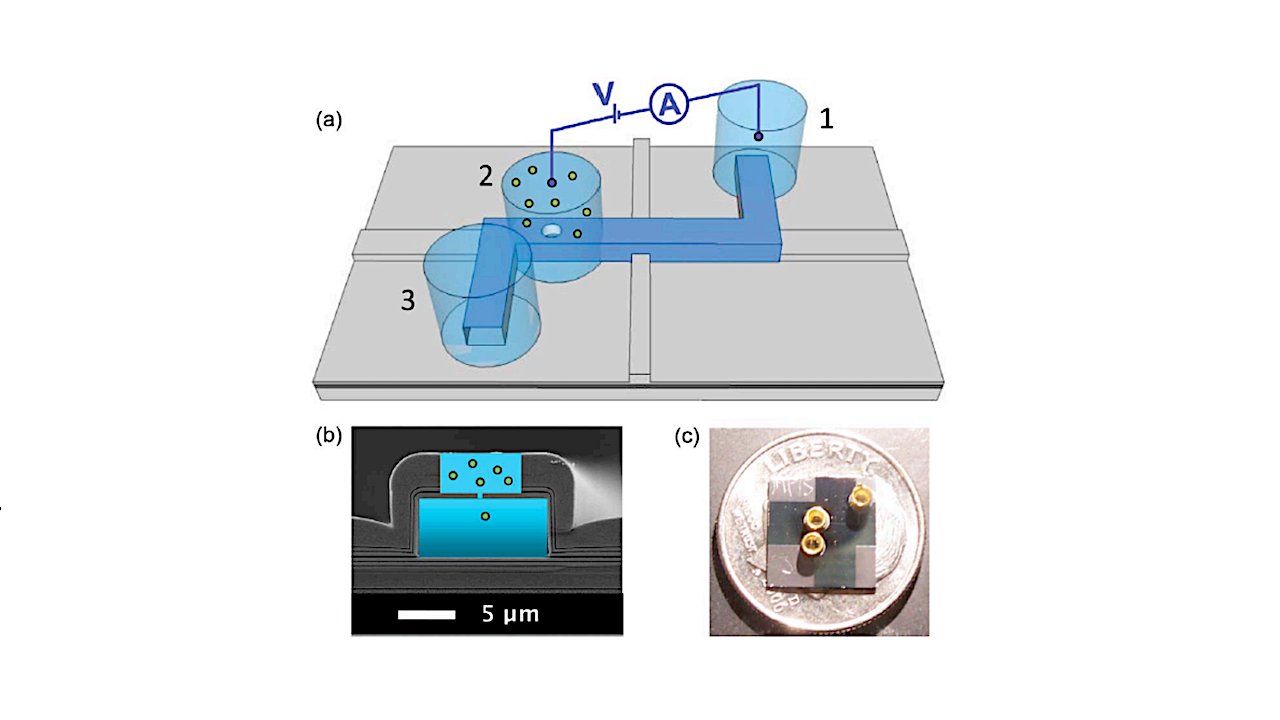
Concept: Ion beam milling and sculpting of silicon oxide and silicon nitride membranes are used to fabricate robust solid-state nanopore membranes with an array of different pore diameters for detection of multiple types of biomarkers (i.e. DNA, charged proteins, amino acids, viruses etc.). Testing parameters (voltage, salinity, pH, etc.) are being established for the detection of biomarkers of interest. We are developing criteria for distinguishing patterns in signal blockades which will indicate the potential for stored information. Optical detection [4], in combination with electrical detection, for multimodal identification is also explored.
DEVELOPMENT OF SOLID-STATE NANOPORE TECHNOLOGY FOR LIFE DETECTION. K. B. Bywaters1 , H. Schmidt2 , W. Vercoutere3 , D. Deamer2 , A.R. Hawkins4 , R. C. Quinn3 , A. S. Burton5 , and C. P. McKay3 , 1 NASA Postdoctoral Program ([email protected]), 2 University of California, Santa Cruz ([email protected]; [email protected]), 3 NASA Ames Research Center ([email protected]; [email protected]; [email protected]), 4 Brigham Young University ([email protected]), 5 NASA Johnson Space Center ([email protected])
References: [1] Benner S. A. and Hutter D. (2002) Bioorganic chemistry, 30, 62–80. [2] Deamer et al. (2016) Nat Biotech 34, 518-24. [3] Castro-Wallace S. L. et al. (2016) bioRxiv:077651. [4] Liu S. et al. (2014) Nano Lett., 14, 4816-4820.
Acknoledgements: This project is funded by the NASA Research Opportunities in Space and Earth Sciences Program’s Concepts for Ocean worlds Life Detection Technology, Solicitation: NNH16ZDA001N-CLDTCH.
Astrobiology