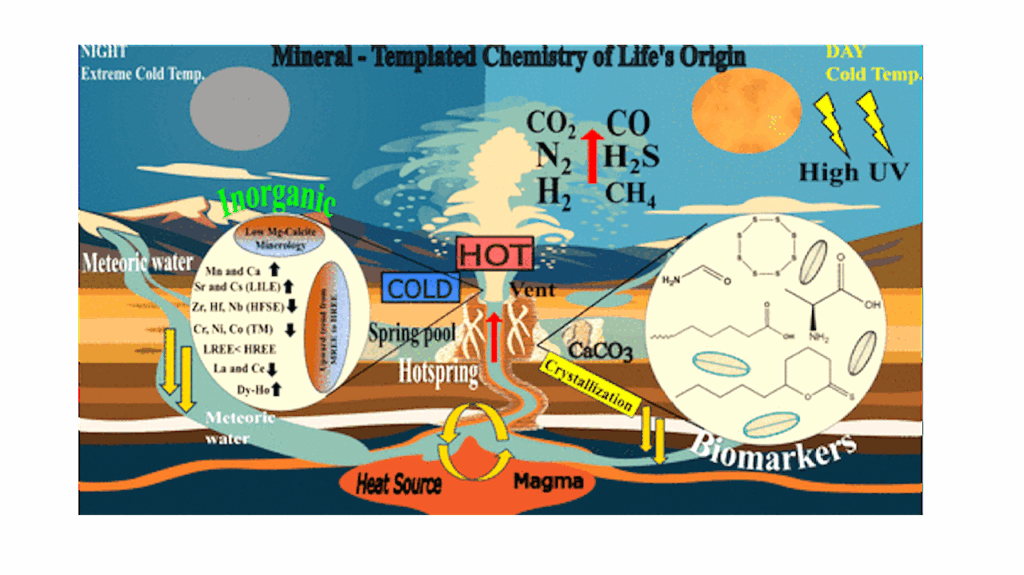
How Life Survives Extreme Environments
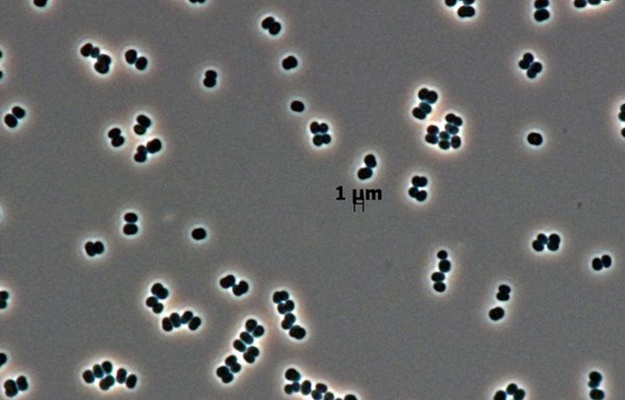
To better understand how microbes behave in extreme environments, one possible proxy, not often considered by astrobiologists, is the human body. Over billions of years of evolution, certain species of microbes inside humans have adapted to environments in the human body that would be extremely rough to many other organisms.
For example, consider a microbe that lives in tissue associated with the immune system, which is designed to attack foreign bodies. The microbe would need to fit a very specific set of biological parameters in order to persist without an attack by the immune system. Also, outside of the gut, most parts of the human body provide few nutrients to sustain microbes living there.
In this respect, Mayo Clinic, a nonprofit organization committed to clinical practice, education, and research, is interested in some of the same origins-of-life studies that drive astrobiologists. Specifically, Mayo researchers want to know how cells pick up cues from their surrounding environment to survive.
Horizontal gene transfer
A group at Mayo, led by Nicholas Chia, an assistant professor of biophysics and surgery, is examining the mechanics of horizontal gene transfer. He first became interested in sequence homology, or how biological sequences are related to each other, while completing his doctorate degree in theoretical biophysics at Ohio State University. Prior to joining Mayo, Chia deepened his understanding of the origins of life with research funded by a predecessor research grant to the current NASA Astrobiology Institute (NAI) for Universal Biology at the University of Illinois at Urbana-Champaign. (The research was then under the Biocomplexity Theme of the Institute for Genomic Biology and funded by the National Science Foundation and the U.S. Department of Energy.)
Chia’s current research is funded by the National Institutes of Health under a grant titled, “Microbial metabolic toxicity drives colon cancer.” His project explores a hypothesis that microbial-producing DNA toxins (like hydrogen sulfide) lead to an increased risk of colorectal cancer. If this hypothesis is correct, sulfate-reducing bacteria in the colon could promote the growth of cancer unless their hydrogen sulfide production is inhibited. Sulfate-reducing bacteria produce hydrogen sulfide, a known genotoxin. It damages DNA that can result in mutations that lead to cancer. Understanding how this process occurs requires studying the complex microbial dynamics within the gut micro-biome. One of these dynamics, horizontal gene transfer, most likely has its origins near the time of the origin of life.
In the animal world, species perform horizontal gene transfer through sexual reproduction, but bacteria achieve the same end through division. This means that bacteria have a different method of mixing their genes; the genes can travel from one microbe directly to another. In many cases, the microbes do not have to come from the same species to achieve a successful transfer, Chia said. “It’s the equivalent of us picking up a dog tail,” he said.
Horizontal gene transfer represents something that many evolutionary theories do not take into account, because evolution mostly talks about change happening gradually. Microbes, by contrast, can pick up new genes as the environment changes around them, even within the space of a few weeks. Should the environment become more acidic, for example, natural selection would favor genes that allow reproduction despite these traits.
“The way this connects to astrobiology is in trying to figure out what are the limits of life,” Chia said. “As it turns out, horizontal gene transfer enables life to exist in more places, and survive in more environments and to evolve faster, than if you just have linear evolution.”
Even more astounding, the microbes can store these genes over time into a sort of field of genes for microbes in that particular environment, known as a pangenome. The pangenome refers to genetic material in the ecosystem. This means organismal and viral genomes. Genes that appear dormant can reside in viruses, dormant bacteria and archaea (single-celled microorganisms), or be within unexpressed sites of a genome waiting to be carried by horizontal gene transfer to a more prominent organism.
A species’ capability could go dormant for a long time, and then be resurrected as the environment requires it. With environments on Mars going through extreme seasonal change, this represents hope that bacteria on this planet could be more resilient than previous theories took into account.
Chia’s research into colorectal cancer is ongoing; one paper describes the first tool for assessing community microbial metabolism based on the standard 16S rDNA backbone (Biorxiv). The team also performed a published survey of the microbial genomes within the human body, yielding three previously uncultivated species from which they sequenced “essentially” complete genomes (Frontiers in Microbiology).
Applications for Mars exploration
Chia’s theoretical biology group includes Marina Walther-Antonio, an assistant professor of surgery. She splits her time working for Mayo and for the Habitability of Martian Environments (HOME) project funded by the European Research Consortium (ERC). The findings of this project are meant to support the European Space Agency (ESA) ExoMars rover project, slated to launch to Mars in 2020.
Like Chia, her research has roots in the NASA Astrobiology Institute, including her participation in the NAI-funded 2003-2008 Indiana-Princeton-Tennessee Astrobiology Initiative to find subsurface microbial communities that could generate bio-sustainable energy and nutrient cycling. Nutrient cycling describes how a nutrient moves through an ecosystem or planet. For example, carbon cycling describes how carbon gets used by life, such as algae at the top of the ocean. The algae then die and become detritus, and as they sink they in turn are eaten and their carbon is utilized by other organisms. Walther-Antonio’s research aims to improve the possibilities of finding and exploring the microbial components of Martian analog environments, such as Warner Valley, Oregon.
At Mayo, Walther-Antonio is working with an optofluidic platform that would allow for analysis of individual cells, rather than cell colonies. (Her research there mostly focuses on women’s health across several pathologies.) The platform uses a laser that relies on using the refractive index difference between a cell and its suspension fluid. A microscope on the platform focuses the light, which is then split into multiple beams to create multiple traps to analyze the cells. This laser tweezer has successfully been used to trap individual cells of Staphylococcus aureus, a common bacteria in the nose and respiratory system. The technology uses a micro-fluidic environment to allow the micro-manipulation of the cells through flow channels and chambers.
“One of the problems we have is that the microenvironment of tumors appear fairly heterogeneous,” Walther-Antonio said. Yet, the picture changes when the tumors are examined on an individual cell level, rather than at the commonly viewed level of a mass of cells, she added. “Different cells can have different genomes, so we can see exactly what is coming from each cell if we can separate them.”
The Mars research — called Habitability of Martian Environments (HOME), of which Walther-Antonio is co-investigator — is funded by the European Research Consortium. The group is using the optofluidics platform to study samples from Chile’s Atacama desert at the single-cell level rather than at a colony cell level. The group conducted two field seasons in 2015 and 2016 to examine a moisture transect in the desert and other specialized habitats that could host microbes able to live in the harsh desert environment. The technology used in this analysis may be promising to study future Martian samples. This would appear to be a more likely prospect for discovering li
fe on the Red Planet, where subzero temperatures and scarce, briny water would present challenging environments for even hardy microbes.
The ExoMars rover site will not target the most prevalent briny water sites, known as recurring slope linae, out of concern for planetary contamination given that active microbes could live there. The four landing sites under consideration likely hosted ancient water and by extension, could have been habitable at some point in the past. Mawrth Vallis and Oxia Planum both have ancient, 3.8-billion-year-old clay rocks that were likely formed in water. Hypanis Vallis is on a fluvial fan, which presumably is the leftover of a river delta from about 3.45 billion years ago. The final site, a channel called Aram Dorsum, provides sedimentary rocks likely deposited through river sediments.
It is anticipated that the technology under development will provide important advancements in our understanding of life at the cellular level both inside and outside our own bodies and planet.
Elizabeth Howell – NASA Astrobiology Institute