A Living Desert Underground

In the perpetual darkness of a limestone cave, UA researchers have discovered a surprisingly diverse ecosystem of microbes eking out a living from not much more than drip water, rock and air
Hidden underneath the hilly grasslands studded with ocotillos and mesquite trees in southeastern Arizona lies a world shrouded in perpetual darkness: Kartchner Caverns, a limestone cave system renowned for its untouched cave formations, sculpted over millennia by groundwater dissolving the bedrock and carving out underground rooms, and passages that attract tourists from all over the world.
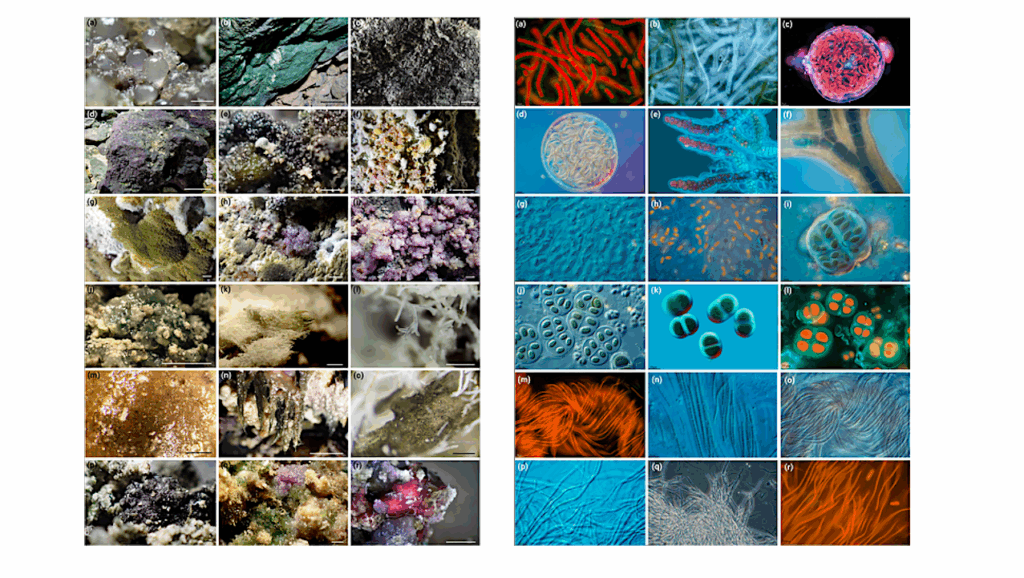
Beyond the reaches of sunlight and seemingly devoid of life, the caves are in fact teeming with an unexpected diversity of microorganisms that rival microbial communities on the earth’s surface, according to a new study led by University of Arizona researchers that has been published in the journal of the International Society for Microbial Ecology. The discovery not only expands our understanding of how microbes manage to colonize every niche on the planet but also could lead to applications ranging from environmental cleanup solutions to drug development.
“We discovered all the major players that make up a typical ecosystem,” said Julie Neilson, an associate research scientist in the UA’s College of Agriculture and Life Sciences. “From producers to consumers, they’re all there, just not visible to the naked eye.”
In a long-standing collaboration between Kartchner Caverns State Park and the UA, Neilson and her co-workers have spent years exploring the underground world and its inhabitants. For their latest study, they swabbed stalactites and other cave formations for DNA analysis. Based on the genes they found in their samples, they reconstructed the bacteria and archaea – single-celled microorganisms that lack a cell nucleus – living in the cave. Kept secret for 14 years after its discovery in 1974 by two UA graduate students who were hiking in the Whetstone Mountains just south of Benson, Ariz., Kartchner Caverns has been protected from human impact so that scientists can study the fragile environment and organisms inside the cave.
“We didn’t expect to find such a thriving ecosystem feasting on the scraps dripping in from the world above,” Neilson said. “What is most interesting is that what we found mirrors the desert above: an extreme environment starved for nutrients, yet flourishing with organisms that have adapted in very unique ways to this type of habitat.”
Unlike their counterparts on the surface, cave microbes can’t harness the energy in sunlight to build organic matter from carbon dioxide in the atmosphere. This process, known as photosynthesis, forms the basis of all life on Earth.
In the absence of light, bacteria live off water runoff dripping into the cave through cracks in the overlying rock and harvest the energy locked in compounds leaching out from decaying organic matter in the soils above and minerals dissolved within the rock fissures, Neilson and her team discovered.
“Kartchner is unique because it is a cave in a desert ecosystem,” Neilson explained. “It’s not like the caves in temperate areas such as in Kentucky or West Virginia, where the surface has forests, rivers and soil with thick organic layers, providing abundant organic carbon. Kartchner has about a thousand times less carbon coming in with the drip water.”
“The cave microbes make a living off the extremely limited nutrients that are available,” Neilson said. “Instead of relying on organic carbon, which is a very scarce resource in the cave, they use the energy in nitrogen-containing compounds like ammonia and nitrite to convert carbon dioxide from the air into biomass.”
The researchers found evidence of cave microbes engaging in all six known pathways that organisms use to fix carbon from the atmosphere to make food and structural material.
Neilson said although the nitrogen-driven pathway is probably the most dominant in the cave, there might be others. Some microbes even eat rock – to derive energy from chemical compounds such as manganese or pyrite.
Raina Maier, a professor in the Department of Soil, Water and Environmental Science at the College of Agriculture and Life Sciences – the study’s principal investigator – said the team expected to find the overall microbial diversity in the cave to be only a fraction of that found in the soil on the surface.
“We expected the surface community many times more diverse than the cave,” said Maier, who is also a member of the UA’s BIO5 Institute. “Instead, we found the cave is about half as diverse as the terrestrial environment where there is sunlight and soil and vegetation. At the same time, the two ecosystems share only 16 percent of the microbial species. In other words, there is a difference of 84 percent between the two, which is amazing.”
Previous studies had shown that, to the cave microbes, the stalactite they live on is like an island: Restricted to the stalactite they happen to be on, there appears to be little mixing between populations, resulting in different assemblages from one cave formation to another.
To analyze the DNA swabbed from the cave formations, Neilson and her team enlisted the help of the lab of Rod Wing, a professor in the UA’s Department of Plant Sciences and director of the Arizona Genomics Institute at BIO5.
“When you work in extreme starving environments, you barely get enough DNA,” Neilson explained. “In some of our samples we got about half of what is considered the minimum amount for DNA sequence analysis. But, we said, let’s just try it. And the wonderful technicians in Rod Wing’s lab tried all those new techniques and they managed to get us a data set even from the dry rock, where there is no drip water and where there are very few microbes living to begin with.”
In addition to encountering all the major players that make up a complex food web in the cave, the scientists discovered what likely are microbes yet unknown to science.
“Twenty percent of the bacteria whose presence we inferred based on the DNA sequences were not similar enough to anything in the database for us to be able to identify them,” Neilson said. “On one stalactite, we found a rare organism in a microbial group called SBR1093 that comprised about 10 percent of the population on that stalactite, but it represented less than 0.5 percent of the microbes on any of the others.”
According to Neilson, nobody has been able to culture that organism in the lab, and its DNA sequence has only ever been found three times in history: in a stromatolite – a special type of sedimentary rock involving microbial communities – in the hypersaline waters of Shark Bay in Australia; in a site contaminated with hydrocarbons in France; and in a sewage treatment plant in Brisbane, Australia.
“This suggests there are many microbes out there in the world that we know almost nothing about,” she said. “The fact that these organisms showed up in contaminated soil could mean they might have potential for applications such as environmental remediation. The most abundant microbe that we found in our taxonomic survey was closely related to a microbe that produces erythromycin, an antibiotic. That is not what it is doing in the cave, but it shows you that not only is there a potential to find microbes that are new to science, but studying them in those extreme and poorly studied environments could lead to new applications.”
The implications of the research reach far beyond Kartchner Caverns, as far as other planets, as the researchers point out.
“There is a lot we have to learn about microbes and how they control processes of global importance, and by studying microbes in extreme ecosystems such as Kartchner Caverns or in the Atacama Desert in Chile, it helps us study some of the capabilities we don’t yet understand in rich ecosystems here on the surface,” Neilson said.
“It shows the flexibility of microbes,” Neilso
n said. “They have conquered every niche on the planet.”
Maier added: “When you think about exploring Mars, for example, and you look at all those clever strategies that microbes have evolved and tweaked over the past 4 billion years, I wouldn’t be surprised if we found them elsewhere if we just keep looking.”
###
Funding for this work was provided by a National Science Foundation (NSF) Microbial Observatory grant and a UA NSF IGERT Genomics Initiative fellowship to first author Marianyoly Ortiz.
Research article: http://www.nature.com/ismej/journal/vaop/ncurrent/abs/ismej2013159a.html